Biology Reference
In-Depth Information
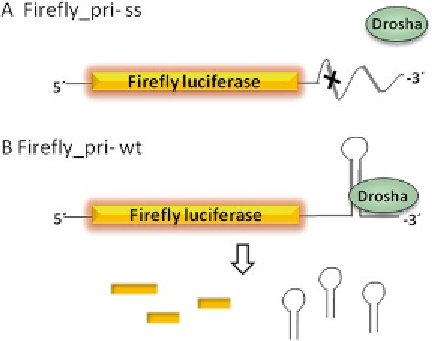
Fig. 1
Schematic representation of luciferase-based measurement of DROSHA
processing activity
III domains and one double-stranded RNA-binding domain. The
most important cofactor is a DiGeorge critical region 8 protein
(DGCR8, also known as Pasha) that forms a microprocessor
complex with DROSHA [
6
].
It is believed that DROSHA does not possess sequence
specificity. Instead it recognizes the RNA secondary structure. Pri-
miRNAs that are recognized by DROSHA have a hairpin structure
with a large, unstructured terminal loop and a stem of more than
26 base pairs (bp) but are less than 40 bp in length [
7
]. DROSHA
preferentially cleaves 11 bp away from free ends of pri-mRNA at
the base of the hairpin stem [
8
].
In our assay we used this recognition feature of DROSHA enzyme
to quantify its enzymatic activity in live cells. As an exemplary pri-miR,
we chose pri-16-1 because its recognition and processing by the micro-
processor complex have been extensively studied. We constructed two
plasmids: one plasmid contains the hairpin fragment of the pri-miRNA
to be analyzed with an additional 25 bp on each side of the hairpin.
This wild-type (wt) sequence is cloned at the 3′UTR of the firefly gene.
Here we used the pri-miRNA-16-1 sequence. The second plasmid is
used as a control plasmid and contains the same sequence (here of pri-
miRNA-16-1) but with four point mutations that were introduced
into the hairpin stem. These mutations destabilize the hairpin struc-
ture. This control plasmid can therefore not be recognized and cleaved
by DROSHA and serves as a positive normalization control for
sequence-dependent and sequence-independent effects of the 3′UTR
(
see
Fig.
1
). Both constructs should have a similar stability [
2
].
2
Materials
Medium should be stored at 4 °C and warmed up to 37 °C
before use.
2.1
Cell Culture