Biomedical Engineering Reference
In-Depth Information
80
50
40
60
30
40
20
20
10
0
0
0
10
20
30
Time (min)
40
50
60
0
10
20
30
Time (min)
40
50
60
A
B
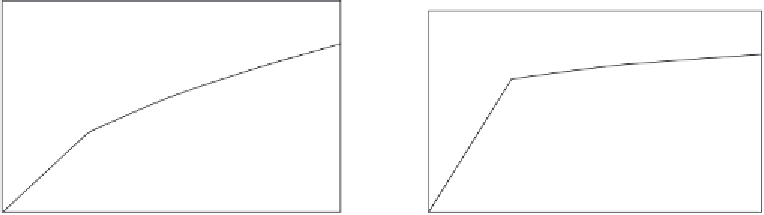
Figure 11.8
Binding (hybridization) of (a) complementary and (b) a noncomplementary (three-base mismatch
strand) DNA in solution to a 30-mer 3
0
-thiolated DNA strand immobilized on an electrochemical
enzymatic genosensor (
Abad-Valle et al., 2007a,b
)
Figure 11.8a
shows the binding (hybridization) of a complementary DNA in solution to a
30-mer 3
0
-thiolated DNA strand immobilized on an electrochemical genosensor (
Abad-Valle
et al., 2007a,b
). A single-fractal analysis is adequate to describe the binding kinetics. The
values of the binding rate coefficient,
k
, and the fractal dimension,
D
f
, for a single-fractal
analysis are given in
Tables 11.4
and
11.5
.
Figure 11.8b
shows the binding (hybridization) of a three-base mismatch DNA strand to a
30-mer 3
0
-thiolated DNA strand immobilized on an electrochemical genosensor (
Abad-Valle
et al., 2007a,b
). Once again a single-fractal analysis is adequate to describe the binding kinetics.
The values of the binding rate coefficient,
k
, and the fractal dimension,
D
f
, for a single-fractal
analysis are given in
Tables 11.4
and
11.5
. It is of interest to note that as one goes from the bind-
ing of the complementary DNA to the three base-mismatch strand in solution to the 30-mer
3
0
-thiolated DNA strand immobilized on the electrochemical genosensor, the fractal dimension
increases by a factor of 1.427 from a value of
D
f
equals to 1.9290 to 2.7520, and the binding rate
coefficient,
k
increases by a factor of 3.353 from a value of
k
equal to 7.0291 to
k
equal to
23.569. Increases in the degree of heterogeneity or the fractal dimension on the sensor chip
surface and in the binding rate coefficient are in the same direction.
Wang et al. (2007
) recently analyzed the binding of complementary ODN (ODN-P)
(2-diolgonucleotide) and a noncomplementary ODN-N (nonmatching) to an electrochemical
sensor with a EST2-A34 reporter. These authors used esterase 2-oligonucleotide conjugate as
a sensitive reporter for the electrochemical detection of nucleic acid hybridization.
Figure 11.9a
shows the binding of
p
-aminophenylbutyrate/esterase 2 from
Alicyclobacillus
acidocaldarius
plus oligonucleotide (ODN) in solution to a site-specific manner ODN-P (per-
fectly matched; complementary) immobilized on an electrochemical biosensor surface.
A dual-fractal analysis is required to adequately describe the binding kinetics. A single-fractal