Biomedical Engineering Reference
In-Depth Information
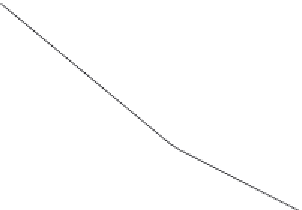
10
9
8
7
6
5
4
2.7
2.75
2.8
2.85
2.9
2.95
Fractal dimension,
D
f2
Figure 9.6
Decrease in the binding rate coefficient, k
2
, with an increase in the fractal dimension, D
f2
.
heterogeneity on the sensor chip surface (in this case in the dissociation phase) leads to an
increase in the dissociation rate coefficient.
Figure 9.6
and
Tables 9.5
and
9.6
show the increase in the binding rate coefficient,
k
2
, with
an increase in the fractal dimension,
D
f2
, for a dual-fractal analysis. For the data shown in
Figure 9.6
, the binding rate coefficient,
k
2
, is given by:
D
10
:
783
2
:
271
k
2
¼ð
466298
63304
Þ
ð
9
:
4
Þ
f2
The fit is very good. Only four data points are available. The availability of more data points
would lead to a more reliable fit. The binding rate coefficient,
k
2
, is extremely sensitive to the
fractal dimension,
D
f2
, or the degree of heterogeneity that exists on the sensor surface as
noted by the greater than negative order of dependence between 10 and a half and 11 (equal
to
10.783) exhibited.
Mank et al. (2006)
recently pointed out that biosensors based on the green fluorescent protein
(GFP) are important tools in cell biology and neuroscience (
Zhang et al., 2002; Miyawaki,
2003; Griesbeck, 2004
).
Mank et al. (2006)
have developed a new generation of calcium
biosensors that use variants of troponin C (TnC) (a specialized calcium sensor of skeletal
and cardiac muscle) as calcium binding moieties. These authors emphasize that their FRET
(fluorescence resonance energy transfer)-based calcium biosensor which includes increased
ion selectivity, moderate calcium sensitivity, and strongly increased maximum fluorescence
is a useful tool for
in vivo
imaging experiments. They emphasize that their biosensor is fast,
and is stable in imaging experiments. Also, their biosensor exhibits enhanced fluorescence
change.
Mank et al. (2006)
report that the most critical test for their biosensor is its
in vivo
perfor-
mance in cells and subcellular compartments of interest. These authors engineered transgenic