Biomedical Engineering Reference
In-Depth Information
O
O
“tether”
-
CH
2
-(CH
2
)
n
N C
H
O
1000
N
H
C
O
N
S
peptide
N
H
where n=
5
O
0
750
-[CH
2
]
n
-
500
where n=
1
3
5
250
0
E. coli
Salmonella
E. coli
Salmonella
Cecropin A
Melittin
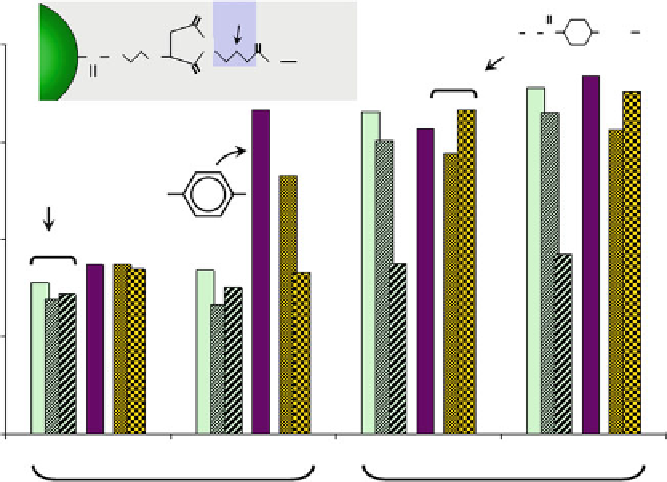
Fig. 5 Effect of “tether” region on binding activity of AMPs immobilized on Luminex
microspheres. Cecropin and melittin were immobilized onto thiol-decorated microspheres using
heterobifunctional (maleimide/
N
-hydroxysuccinimidyl ester) cross-linkers with different “tether”
regions (shown)
rapid, and broad spectrum the sensor, the better suited it will be for on-site use.
Integration of AMPs into biosensors has the potential to expand the number of
bacterial, viral, and fungal targets detectable in a single test, while also providing a
more heat-, solvent-, pH-, and salt-stable biorecognition species than most
antibodies currently used in biosensors.
QCM and SPR-based systems utilizing AMPs have the advantage of
“reagentless” detection; binding of microbial targets can be directly detected,
without the need for a labeled species. Systems such as those described by Mannoor
[
41
] and Mello [
43
,
51
] therefore offer tremendous potential for such broad
spectrum, rapid microbial detection. Unfortunately, instrumentation for these and
other reagentless platforms has generally not yet evolved with sufficient robustness
for routine use outside of a highly controlled, pristine environment, and their
multiplexing capacities are still limited.
Biosensors intended for outside-the-lab use, on the other hand, often utilize
electrochemical, optical, or other tags that can be used to discriminate detected
target from external sources of signal. These biosensors therefore most often
required use of labeled “tracer” species (e.g., antibody, AMP, alternative ligand)
for target detection. To take full advantage of the broad specificity of AMP-based
biosensors, this tracer should be capable of tagging all bound targets equally, such
Search WWH ::

Custom Search