Biomedical Engineering Reference
In-Depth Information
crystal structure to be obtained from x-ray diffraction data. The diffrac-
tion peaks can be used to reconstruct an electron density map, which
is accomplished by Fourier transforming the diffraction intensities. Ob-
viously, analysis of x-ray diffraction data is a computationally intense
exercise!
X-ray crystallography is only useful if crystals can be grown from
purified proteins. This is because the presence of precise, repeating
structure present in crystals is essential for using x-ray diffraction to
solve 3-dimenstional structures. For this reason, the structure of many
intact proteins cannot be accomplished using x-ray diffraction. Conse-
quently, a “divide and conquer” approach is often used, in which protein
are expressed as individual domains for structural analysis. A protein do-
main is typically defined as an independent folding unit within an overall
protein structure. Thus, proteins can be broken down into their compo-
nent parts. Individual domains of interest, such as regions that contain
catalytic sites of enzymes or protein interaction domains, are crystallized
and the structure of these domains independently solved.
The resolution of the structure obtained with x-ray diffraction is highly
dependent on the quality of the crystals that can be grown, Better crys-
tals can usually be produced with smaller molecules (or protein domains)
than with larger molecules. The structural resolution is defined in terms
of angstroms (Å): for example, the crystal structure of protein X at 2.4 Å,
or 1.8 Å, etc. An Å is 0.1 nanometer, or meter. At 2 Å resolution,
peptides can distinguished in a protein, but resolutions of 1.5 Å or better
are required to distinguish individual atoms, which allows much more
precise understanding of the molecule. An clear understanding of the
3-dimensional structure of a protein (or protein domains) is crucial for
a variety of studies, including mapping contact sites between an en-
zyme and its substrate, delineating the nature of antigen binding sites
in antibodies or identifying potential target sites for therapeutics in the
treatment of viral infections. High resolution structural analysis is also
important for helping design agonistic or antagonistic ligands for recep-
tors that function abnormally in disease, to name but a few of the many
ways structural information has been useful.
Nuclear magnetic resonance (NMR) spectroscopy
For those proteins that cannot be crystallized, NMR is the method of
choice for tertiary structural analysis. NMR was first developed in the
1940s but it wasn't until the 1970s and later that it could be applied to
solving structures of biomolecules. While NMR is generally less sensitive
than x-ray diffraction and data collection requires longer periods of time,
recent advances have significantly improved NMR technology.
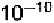
Search WWH ::

Custom Search