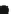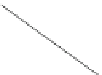Biomedical Engineering Reference
In-Depth Information
$s
s
a
b
c
g
a
1
g
a
2
a
a
$a$
a
g
a
10
b
$b$
b
s
b
a
c
c
c
c
b
s
c
a
b
c
a
b
Fig. 3.1
Circuit construction example:
a
predictor set shown as connectivity graph and
b
function
enumeration and MUX for gene
a
shown in detail
3.3.1.3
Example
To illustrate the method, we consider a small 3 gene example. Let us label the genes
in our example Boolean network as genes
a
,
b
, and
c
, with the following gene logic
functions:
a
=
b
+
c
b
=
a c
c
=
a
+
b
In the notation,
a
,
b
,
c
are the next state variables of
a
,
b
,
c
respectively. From these
functions, we can derive a state transition table (Table
3.1
) which describes the next
state in the BN given a present state. The predictors for each gene of our example are
shown in Fig.
3.1
a), and listed as follows:
p
a
={
b
,
c
}
,
p
b
={
a
,
c
}
, and
p
c
={
a
,
b
}
which predicts (regu-
lates) the activity of gene
i
. Note that in our problem, the logic functions for
a
,
b
,
and
c
are unknown, and are to be determined from gene expression observations.
The gene expression observations that are provided to the GRN determination engine
are any subset of the 8 rows of Table
3.1
.
A predictor
p
i
={
j
,
k
,
..
}
lists the set of genes
{
j
,
k
,
..
}