Biomedical Engineering Reference
In-Depth Information
interparticle repulsive forces, interparticle crosslinking initiates
aggregation of the NPs. Typical biological recognition events
include DNA hybridization; aptamer-target, antibody-antigen,
streptavidin-biotin, and lectin-sugar interactions; and metal-ligand
coordination. Furthermore, biologically relevant organic molecules
(such as peptides) containing residues, such as thiol and guanidine,
can directly induce crosslinking aggregation of Au NPs by chemical
interactions (e.g., Au-S).
3.4.1.1
Protein assays
Crosslinking-based colorimetric assays have been applied to the
determination of a number of target proteins, including streptavidin,
concanavalin (Con A), protein A, and PDGF. In these assays, recognition
molecules are anchored to the Au NP directly or through coupling
reactions as shown in Fig. 3.2. Common recognition molecules for
proteins include biotin, mannose, aptamers, antibodies, and others.
Once speciic interactions between the recognition molecules and
target proteins occur, the aggregation of Au NPs leads to color changes.
Representative examples of crosslinking-based assays using Au NPs
are for the determination of human chorionic gonadotrophin in urine
and serum.
68-70
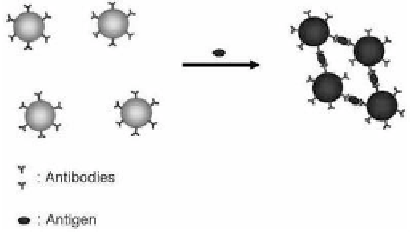
Au NPs conjugated to antibodies are crosslinked
(agglutinated) by multivalent binding to the target molecules
(see Fig. 3.4). In the aggregates, the distances between Au NPs are
~25 nm, which is smaller than the size (60 nm) of the Au NPs
themselves. At high concentrations (>0.1 nM) of target molecule, there
is a visible color change from red to blue, but at lower concentrations,
this change can only be detected with a spectrophotometer.
Figure 3.4
A schematic diagram of a distance-dependent sandwich assay
for high-molecular-weight polyvalent antigen (green ovals)
leading to agglutination of Au NPs and a redshift in their
extinction spectrum. See also Color Insert.
















Search WWH ::

Custom Search