Biology Reference
In-Depth Information
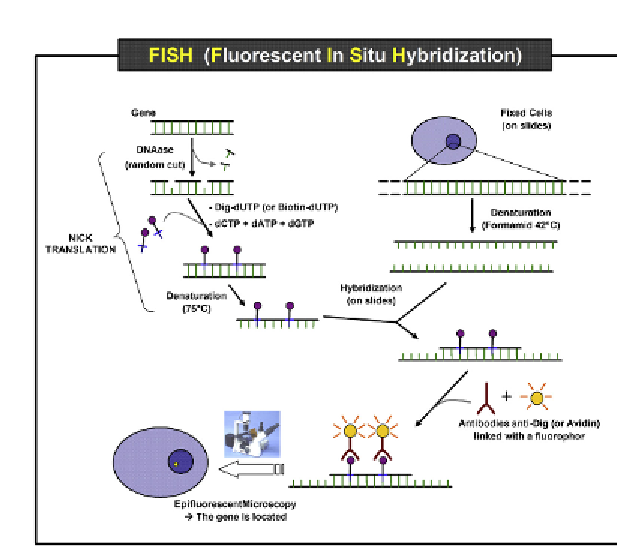
Figure 8.5
Cells from a sample can be fixed on a slide. A probe that will hybridize to a
specific target DNA sequence can be labeled with a fluorophore. The labeled probe and
the target DNA are then denatured and allowed to reanneal. The labeled probes can be
visualized under a microscope and the location (cell) harboring the sequence can be
detected. Source:
Image taken from
http://www.biovisible.com/indexRD.php?page=ish
.
(For color version of this igure, the reader is referred to the online version of this topic.)
8.2.3.2. Polymerase chain reaction
PCR is a technique that allows specific sequences of DNA to be synthesized
over and over again to produce many copies of the same sequence.
43
Although
one copy would be hard to detect among so many other DNA sequences,
many copies of the same sequence can be detected. The amplification process
is possible by using the ability of DNA polymerase to synthesize new strands
of DNA complementary to the offered template strand. Because DNA poly-
merase can add a nucleotide only onto a preexisting 3′-OH group, it requires a
primer. This primer can be complementary to a portion of the target sequence
or to a portion of the DNA adjacent to the target sequence, to which it can
add the first nucleotide. If two primers flank the target sequence, it is possible
to delineate a specific region of template sequence that the researcher wants
to amplify. The sequence can be made over and over again in each round of
replication, which will be accumulated in millions of copies (amplicons) at the
end of the PCR reaction (
Fig. 8.6
).
Search WWH ::

Custom Search