Biology Reference
In-Depth Information
proteins is generally performed at the protein level, whereas mass
spectrometry-based quantifi cation usually occurs at the peptide
level. Proteome analysis based on 2D polyacrylamide gel electro-
phoresis (PAGE) has been substantially improved by the introduc-
tion of 2D DIGE (
1
) with respect to reliability, accuracy, dynamic
range, and reproducibility of protein spot quantifi cation. In 2D
DIGE approaches, fl uorophores are covalently attached to an
amino acid side chain group before electrophoretic separation.
In the frequently applied 2D DIGE minimal labeling concept,
three different CyDyes (Cy5, Cy3, and Cy2) are available, which
are balanced with respect to charge and attached to the
-amino
group of lysines and free N-terminal residues. Typically, only a few
percent of the molecules from each protein species are labeled, and
on each 2D gel, around 50
ε
g of protein sample per CyDye are
co-separated. In 2003, an extremely sensitive modifi cation of the
DIGE concept was developed (
2
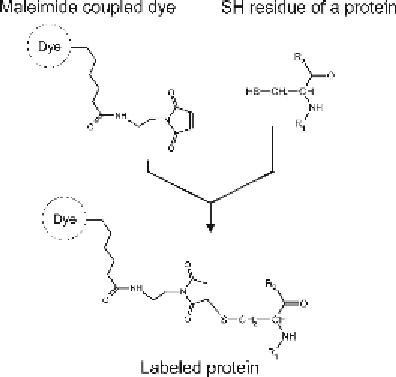
). Since all accessible sulfhydryl
residues of cysteines are labeled to completion in this modifi cation,
it is referred to as “saturation labeling.” Two CyDyes (Cy3 and
Cy5) are available for this system, which are coupled via a maleimide
linker to sulfhydryl groups after chemical protein reduction (see
Fig.
1
). The total protein amount required for 2D gel electropho-
resis of complex cell lysates could thereby be reduced by two orders
of magnitude down to the lower microgram range. This high
sensitivity has made accessible many new areas in biomedicine and
biology to quantitative 2D gel-based proteomic studies, e.g., analysis
of samples from microdissections (
3, 4
), glomerular cell preparations
(
5
), membrane protein preparations (
μ
6
), or mammalian oocytes (
7
).
Fig. 1. Maleimide-based coupling of a CyDye to a cysteine residue.
Search WWH ::

Custom Search