Agriculture Reference
In-Depth Information
Table 9.3
Chemical and physical parameters of PLD alpha proteins encoded by genes in tomato (LePLD
α
1,
LePLD
α
2, and LePLD
α
3), strawberry (FaPLD
α
1), honeydew melon (CmPLD
α
1 and CmPLD
α
2), and
cucumber (CsPLD
α
1)
Amino
MM
ASP
+
ARG
+
Acidic/
Theoretical
Grand average
acids
(kDa)
GLU
LYS
basic
pI
hydropathicity
LePLD
α
1
809
92.2
121
89
A
5.39
−
0
.
447
LePLD
α
2
807
92.0
112
86
A
5.59
−
0
.
349
A
−
LePLD
α
3
806
92.7
105
94
6.29
−
0
.
378
α
−
.
FaPLD
1
810
91.4
114
91
A
5.76
0
395
α
−
.
CmPLD
1
808
92.0
122
92
A
5.37
0
483
A
−
CmPLD
α
2
807
92.1
109
97
6.20
−
0
.
398
CsPLD
α
1
808
91.8
122
92
A
5.37
−
0
.
472
Parameters shown in columns 1-7 from left to right include total encoded amino acids, molecular mass (MM),
sum of aspartic plus glutamic acid residues (ASP
+
GLU), sum of arginine plus lysine residues (ARG
+
LYS),
acidic or basic polypeptide (A, acidic; A
−
, weakly acidic), theoretical isoelectric point (pI), and grand average
hydropathicity. PLD alpha protein accession numbers are shown in Table 9.2.
phospholipase D alpha 2 (CmPLDa2) [Cucumis melo var. inodorus]
phospholipase D alpha 2 (LePLDa2) [Lycopersicon esculentum]
phospholipase D alpha 3 (LePLDa3) [Lycopersicon esculentum]
phospholipase D1 [Craterostigma plantagineum]
phospholipase D2 [Craterostigma plantagineum]
phospholipase D alpha [Cynara cardunculus]
phospholipase D alpha 1 (PLD alpha 1) [Nicotiana tabacum]
phospholipase D alpha 1 (LePLDa1) [Lycopersicon esculentum]
phospholipase D alpha 1 (PLD alpha 1) [Pimpinella brachycarpa]
phospholipase D alpha [Vitis vinifera]
phospholipase D alpha 2 [Arachis hypogaea]
phospholipase D alpha 1 [Medicago truncatula]
phospholipase D alpha 2 [Medicago truncatula]
phospholipase D alpha 1 (CmPLDa1) [Cucumis melo var. inodorus]
phospholipase D [Cucumis sativus]
phospholipase D alpha [Gossypium hirsutum]
phospholipase D alpha 1 (PLD alpha 1) [Vigna unguiculata]
phospholipase D alpha 2 (PLD 2) [Brassica oleracea]
phospholipase D alpha 1 (PLD 1) [Brassica oleracea]
phospholipase D alpha 1 [Arabidopsis thaliana]
phospholipase D alpha [Arabidopsis thaliana]
phospholipase D Alpha 2 [Arabidopsis thaliana]
phospholipase D alpha 1 precursor (PLD 1) [Ricinus communis]
phospholipase D alpha [Fragaria
×
ananassa]
phospholipase D2 [Papaver somniferum]
phospholipase D1 [Papaver somniferum]
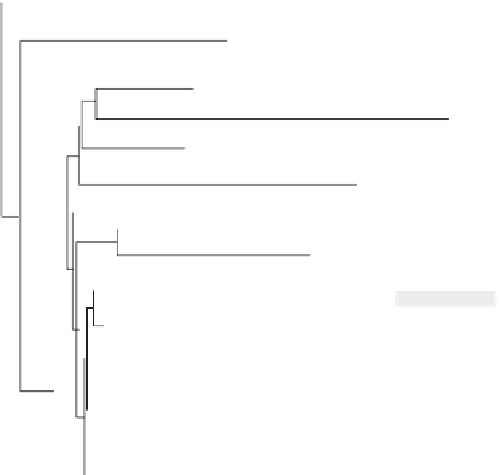
Fig. 9.11
Phylogenetic tree showing the relatedness of plant PLD alphas from 17 dicot species based on alignment
of their complete deduced amino acid sequences ranging from 806 to 813 residues. Accessions with arrowheads
on the right indicate the seven PLD alpha genes currently being investigated in relation to ripening and senescence
of tomato (
Lycopersicon esculentum
—light gray), honeydew melon (
C
.
melo
—medium gray), and strawberry
(
F
.
ananassa
—dark gray) fruits. The other two highlighted accessions indicate PLD alphas cloned from tissues
of grape berry (
V
.
vinifera
—dark gray) and cucumber fruit (
C
.
sativus
—light gray).
×














































































Search WWH ::

Custom Search