Biology Reference
In-Depth Information
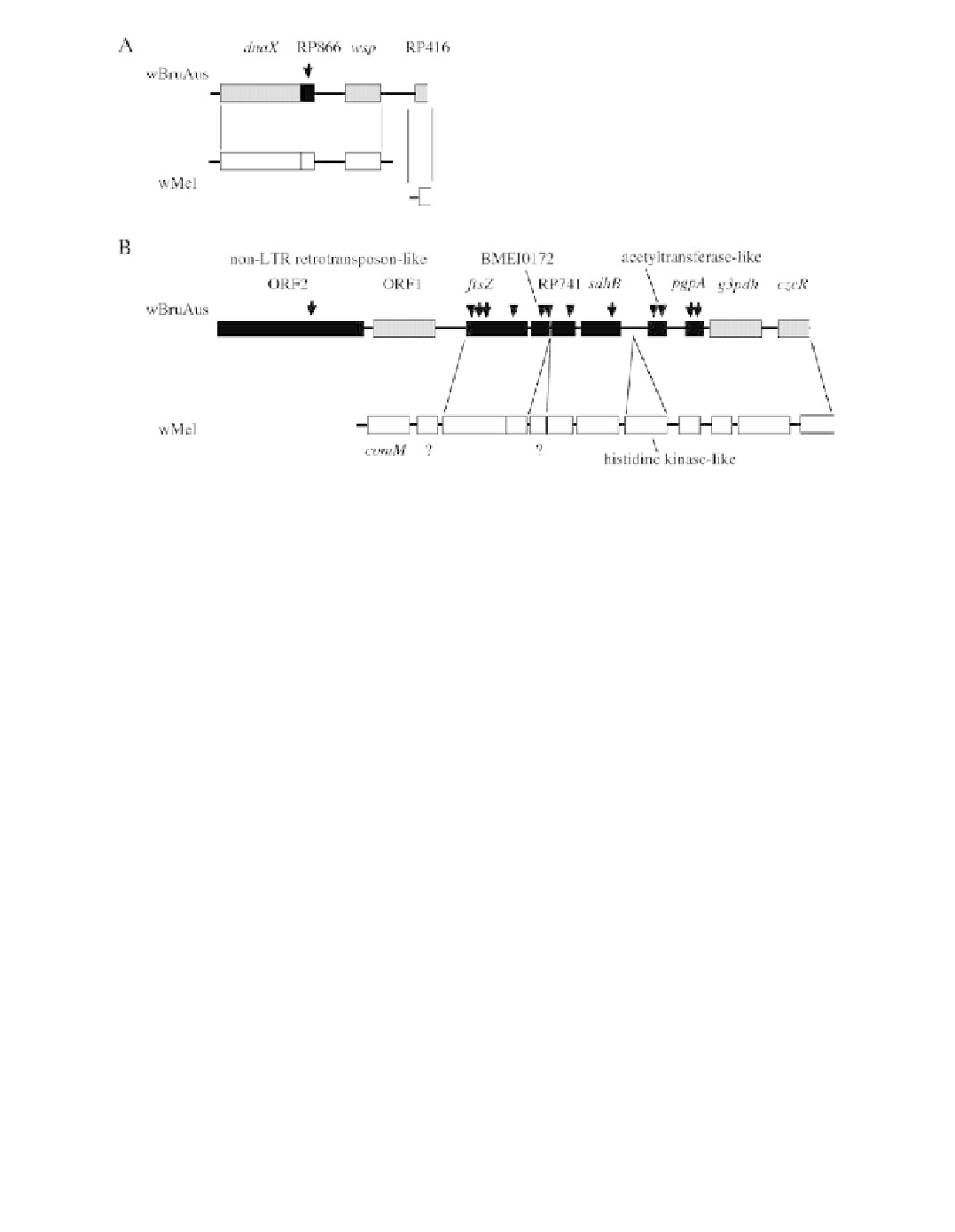
FIGURE 18.12
Structure of the genome fragments of wBruAus obtained by inverse PCR, aligned with
genome sequences of wMel, a strain of
Wolbachia
from
D. melanogaster
. (A) A 4.1-kb fragment containing
wsp
gene. (B) An 11.4-kb fragment containing
ftsZ
gene. Arrows indicate the position of intermittent stop
codons. Arrowheads show the position of frame shift substitutions. Filled ORFs contain either stop codons or
frame shift substitutions, whereas shaded ORFs are structually intact. A non-LTR retrotransposon-like sequence
was located at the upstream of
ftsZ
gene. All ORFs on wMel genome fragments are structurally intact. Question
marks indicate unidentiÝed ORFs. [From Kondo, N., Nikoh, N., Ijichi, N., Shimada, M., and Fukatsu, T.
(2002b).
Proc. Natl. Acad. Sci. U.S.A.
99:
14280Ï14285.]
MosquI
elements from
Aedes aegypti
(Tu and Hill, 1999), was identiÝed in the upstream region of
ftsZ
(Figure 18.12B). The sequence contained two ORFs typical of non-LTR retrotransposons,
ORF1 encoding a protein of unknown function, and ORF2 encoding a protein with endonuclease
and reverse transcriptase activities. The
I
element is known to be involved in the
IÏR
system of
hybrid dysgenesis in
D. melanogaster
(Bucheton et al., 1984). A number of
I
and
Yo u
elements,
which constitute a distinct clade in the non-LTR retrotransposon phylogeny (Berezikov et al., 2000),
have been identiÝed in the genomes of
D. melanogaster
and other Þy species. Since non-LTR
retrotransposons of this family have been found in insect genomes but not in bacterial genomes,
this Ýnding may favor the idea that the
Wolbachia
genome fragment is located on a chromosome
of the host insect.
COMPARISON WITH THE
WOLBACHIA
GENOME
FROM
D. MELANOGASTER
Complete genome sequencing of wMel, a strain of
Wolbachia
from
D. melanogaster
, is now in
progress by the Institute for Genomic Research. Since the preliminary genome sequences are
available through the Web site
(http://www.tigr.org)
,
we compared the genome fragments of wBru-
Aus with those of wMel (Figure 18.12). Gene arrangements on the genome fragments were, in
general, conserved between wBruAus and wMel, although several differences were identiÝed. In
wBruAus,
wsp
was next to RP416, whereas in wMel these genes were not on the same genome
fragment. In wMel only, a histidine kinase-like sequence was found between
sdhB
and acetyltrans-
ferase-like sequence, and an unidentiÝed ORF was located between BMEI0172 and RP741. In the