Biomedical Engineering Reference
In-Depth Information
8.1.1 Biopolymers
Classically, protein expression is described by the following sequence: the genetic
information carried on the DNA sequence is read by a protein assembly called RNA
polymerase; this transcription gives rise to the RNA molecules. The messenger RNA
finds its way out of the nucleus in the cytoplasm and is translated into functional
proteins by the ribosome. We now describe a few elements on the structure of these
macromolecules.
8.1.1.1 DNA Molecules
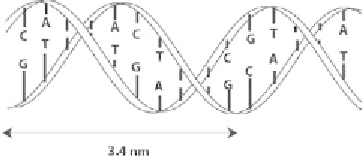
DNA (desoxyribonucleic acid) molecules are made of two strands twisted around
each other. Each of these strands consists of four bases: adenine ([A]), thymine ([T]),
guanine ([G]), and cytosine ([C]) on a phosphate backbone and they are arranged
in a double helix where the bases are located inside and paired exclusively [A]-[T]
and [G]-[C]; they are called Watson-Crick base pairs. The two strands are oriented
and arranged in antiparallel directions (Figure 8.1). The arrangement of the base
pairs along the strand bears the genome of an individual and contains all his or her
genetic information.
Above a certain denaturation temperature, the two strands separate. This prop-
erty is used in the polymerase chain reaction (PCR) technique to amplify the number
of copies of DNA molecules in a given sample. In this technique, after the denatur-
ation step, the sample is cooled down and “primers” that are short complementary
sequences bind to the beginning and end of the region of the DNA to be amplified.
An enzyme (the polymerase) then reads the single strand and matches it with its
complementary sequence using the free nucleotides in solution. So, starting from one
double strand, we end up with two. The same process is cycled 30 to 40 times, lead-
ing to an exponential amplification of the number of copies of the initial sample.
At a much larger scale, DNA is a polymer. When sufficiently diluted, DNA
chains in solution adopt a coil configuration whose radius, called the radius of
gyration
R
g
,
is directly related to the size of the monomers
b
and their number
N
through the relation [1] (Figure 8.2):
b N
ν
R
= ×
(8.1)
For polymers in “good solvent” (meaning that the interactions between a
monomer and a solvent molecule are favored compared with interactions between
two monomers), this exponent
ν
is 3/5. In some cases however, these interactions
are effectively comparable and the chain is said to be ideal, the exponent
ν
is then
Figure 8.1
Double helix structure of a DNA molecule