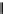Biology Reference
In-Depth Information
The
E. coli Mfd
is a monomeric 130 kDa protein containing eight structural
domains
27
(
Fig. 2
). The N-terminal portion, consisting of D1a, D2, D1b, and
D3, is joined to the C-terminal portion (D4, D5, D6, and D7) by a flexible
linker of 23 amino acids. Although probably not normally associated with
the EC,
29
Mfd
can interact with the
subunit of the RNA polymerase when
the EC is arrested by a DNA lesion, a DNA-bound protein, or by nucleotide
depletion.
16
The
Mfd
protein appears to approach the EC from the upstream
direction, the interaction being inhibited by a
b
subunit
of the polymerase but not by a downstream protein bound to the DNA. If the
EC encounters a pause site, backtracks a few nucleotides, and becomes tran-
siently arrested,
Mfd
can reactivate it and allow it to continue transcribing if
the needed nucleoside triphosphates are present. It apparently does this by
pushing the polymerase forward to realign the RNA transcript in the active site.
Similarly, if the EC has been arrested by nucleotide depletion
Mfd
can facilitate
its restart if nucleotides become available.
16,28,30-32
In contrast, if nucleotides
are not available or if the EC is blocked by a DNA lesion such as a CPD, or a
protein bound to the DNA,
Mfd
can displace the RNA polymerase and the
nascent transcript from the DNA. It has been reported that
Mfd
can also
displace an EC that encounters a head-on replisome complex,
33,34
although
an alternative result has also been described.
35
Studies of the structure and function of
Mfd
have shown that the enzymatic
activities of the purified wild-type protein in solution are inhibited by an
interaction between two of the protein domains, the N-terminal D2 and the
C-terminal D7.
36
This interaction interferes with the RNA polymerase dis-
placement, ATPase, and DNA translocase activities. Changes in the protein
that prevent the interaction of D2 and D7 not only enhance these enzymatic
activities but also result in a protein that is more sensitive to proteases,
indicating significant changes in conformation.
37-40
Presumably, the conforma-
tion of the protein is regulated
in vivo
so that
Mfd
only displaces RNA
polymerase from the DNA when the polymerase is arrested.
s
factor bound to the
b
UvrB homology module
Translocation module
RID
I Ia
II
III
IV
V VI TRG
100
200
300
400
500
600
700
800
900
1000
1100
D1a
D2
D1b
D3
D4
D5 (TD1)
D6 (TD2)
D7
F
IG
. 2. Schematic linear representation of the
Mfd
protein (adapted from Ref.
28
): The UvrA-
interacting domain (Domain 2) is homologous to the N-terminal UvrA-binding domain of UvrB.
The RNA polymerase interacting domain (RID) is shown in pale blue. The translocation domain of
Mfd
, shown in green, contains seven SF2 helicase motifs, indicated with Roman numerals.
The TRG motif, shown in purple, is homologous to the translocase domain of RecG. Numbers
below the bars refer to amino acids.