Biology Reference
In-Depth Information
F
IG
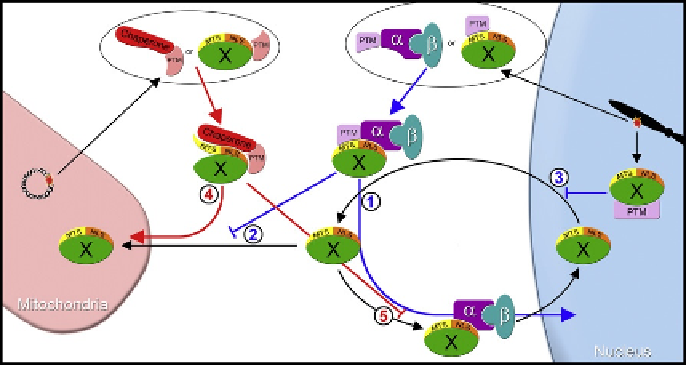
. 2. Dynamic localization of base excision repair (BER) proteins requires localization
signals. Protein X is any BER protein able to localize to both nuclei and mitochondria. Under
nonstress conditions, protein X localizes to both nuclei and mitochondria (black arrows). Under
nuclear or mitochondrial stress conditions, nuclear oxidative DNA damage signals (NODDS) or
mitochondrial oxidative DNA damage signals (MODDS) are generated respectively, resulting in
posttranslational modification (PTM) to either BER proteins or to transport machinery. NODDS
lead to an increased nuclear concentration of BER proteins through one of three mechanisms:
(1) increasing nuclear import through PTM of either protein X or the nuclear import machinery,
enhancing the association of importin
a
/
b
with the classical nuclear localization signal (NLS) of
protein X; (2) decreasing mitochondrial import by PTMs, which directly or indirectly disrupt the
mitochondria matrix-targeting sequence (MTS); or (3) decreasing nuclear export by PTMs that
block a nuclear export signal or create a nuclear retention signal (blue lines). MODDS lead to
increased mitochondrial BER protein concentration through one of two mechanisms: (1) PTMs
causing chaperones to associate with protein X and facilitating mitochondrial import by maintaining
protein X in an unfolded state and enabling mitochondrial import state or (2) decreasing nuclear
import through PTMs, which directly disrupts importin
a
/
b
binding to the NLS of protein X (red
lines).
One of the intriguing features of the
S. cerevisiae
BER system is that most
of the early pathway proteins (glycosylases, AP lyases, AP endonucleases) are
shared by the nucleus and mitochondria, making budding yeast an ideal system
to interrogate the mechanisms regulating BER protein levels, activity,
and localization.
195
The proteins shared by nuclei and mitochondria include
Ntg1, the uracil DNA glycosylase (Ung1), the 8-oxoguanine DNA glycosylase/
AP lyase (Ogg1), and the AP endonuclease (Apn1). In contrast, Ntg2, the
3-methyladenine DNA glycosylase (Mag1) and the AP endonuclease (Apn2)
are strictly nuclear.
34,39,42
Ntg1/2 and Apn1/2 are the result of a genome
duplication in the recent evolutionary past of
S. cerevisiae
, and as such, each