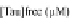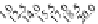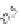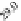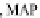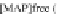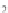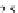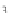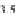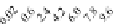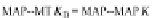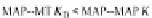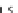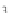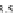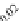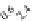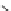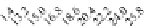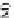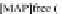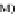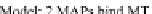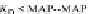Biology Reference
In-Depth Information
B
A
µ
D
C
µ
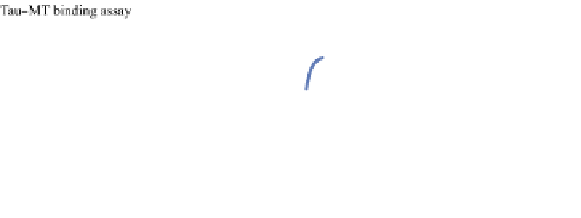
FIGURE 23.2
Tau-MT-binding assay and simulations from MTBindingSim. (A) Data from a Tau-MT
cosedimentation binding assay. (B) MTBindingSim simulation of a first-order binding
relationship with a
M. (C) MTBindingSim simulation using the MAPs bind
MT-bound MAPs model, with the MAP-MAP
K
D
of 0.1
m
K
D
less than (green line), equal to (red line), and
greater than (blue line) the MAP-MT
K
D
. In interpreting these curves, it is important to
remember that a smaller
K
D
value corresponds to a higher affinity. (D) MTBindingSim
simulation using the two MAPs bind MT-bound MAPs model with the MAP-MAP
K
D
less than
(orange line), equal to (teal line), and greater than (purple line) the MAP-MT
K
D
.
Panel A: Reproduced from
Duan and Goodson (2012)
bind the first, already MT-bound, Tau protein. This would result in Tau oligomers
forming on the MT surface (approximately 2-3 Tau proteins binding each other
on the MT surface). Both Tau-MT and Tau-Tau binding would contribute to the
amount of Tau that is measured in the pellet fraction of a cosedimentation assay,
which would normally be interpreted as Tau bound to MTs.
Could this type of model explain the data? Intuitively, it is appealing. MTBin-
dingSim can be used to investigate this question in a more quantitative way. To start,
it is a good idea to investigate how proteins interact according to a simple binding
model. To do this, the standard “first-order binding” model is employed, in which
Tau binding only to MTs is simulated. For the curve shown in
Fig. 23.2
B, the
K
D
was set to 0.1
M based on reasonable estimates from other Tau-MT-binding exper-
iments (data not shown). As shown in
Fig. 23.2
B, the amount of Tau that binds to the
MTs saturates at 1
m
M.
m