Biomedical Engineering Reference
In-Depth Information
A1- 0
A3-0
D1
D3
D5
W1
D2
D4
A1-1
A2-0
A1-1
A1- 0
E1
+
W1
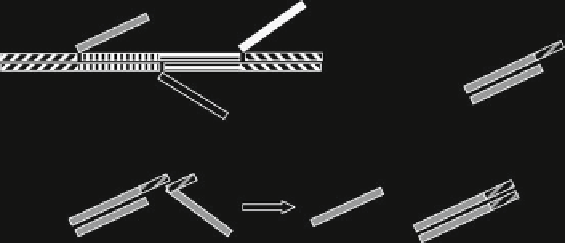
Fig. 7.7
Biomolecular memory.
To p l e f t
addressable branches,
top right
writing procedure,
bottom
erasing procedure
of sites spaced at 7 nm in which specific DNA strands control the bit state of each
address (
Shin and Pierce 2004
). The readout of each state can be performed by
fluorescent molecules that emit at different visible wavelengths. The biomolecular
memory is formed from five single-stranded oligonucleotides denoted by Di in
Fig.
7.7
, which self-assemble in a structure with three addressable branches Ai if
stoichiometrically mixed.
The branches Ai, which are separated by 20 bp with a total length of 7 nm,
have the logic value 0 if single stranded and the logic value 1 if double stranded.
The information can be written by single-stranded writing molecules Wi, which
have a complementary structure to one of the addresses, changing its logic values
from 0 to 1 by hybridization. The writing strands have a 10-bp overhang, which
represents the nucleation site for hybridization with a matching region of an erasing
strand. The erasing strands Ei restore the initial 0 logic value of the address by
hybridizing with Wi and thus forming a duplex waste product Wi and a single-
stranded address with a 0 logic value. The state readout can be done by fluorescent
measurements if quencher molecules for different fluorescent molecules in the
vicinity of the addresses are attached to the Wi strands. Then, the 0 state is
associated to no (or weak) fluorescence, and the 1 logic state corresponds to
strong fluorescence. The writing and erasure processes must be implemented in a
controlled sequence to avoid undesirable interactions between strands. This memory
device is slow: the writing rate constants are of the order of 10
5
s
1
M
1
, while the
erasing process is governed by a fast phase with a comparable rate to the writing
process and a slow phase, with a rate of 10
3
s
1
M
1
. Larger memory addresses
require more rigid, two-dimensional scaffolds.
Biomolecules can also be used to implement a fractal memory, which is no longer
a binary-code device, but its output signal pattern depends on the pattern of the
input signal. For example, for input pulses of different shapes, such as in Fig.
7.8
,
the output represented, for example, by the decay of a photocurrent is different.
Different patterns of the input signal can represent different ASCII codes.
Search WWH ::

Custom Search