Biomedical Engineering Reference
In-Depth Information
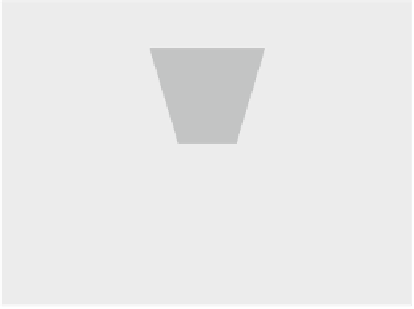
Fig. 3.11
DNA stretch with
optical tweezers
z
x
bead
trap center
bead
x
ds DNA
z
bead
coverglass
light
RNA polymerase
end of the DNA on a cover glass with the help of stalled RNA polymerase, while
the other end was linked to a bead trapped in the optical tweezer. By moving the
cover glass and recording the bead with nanometer resolution with the help of an
interferometric detector (see Fig.
3.11
), the double-clamped dsDNA is stretched and
the force dependence on the distance x can be determined (
Wang et al. 1997
).
If the stiffness k
x
at the trap center is given by
k
x
˝
x
bead
˛
D
k
B
T; (3.38)
where
˝
x
bead
˛
is the mean-square thermal displacement of the bead, the force
dependence on distance is given by
k
x
x
bead
cos
h
tan
1
z
trap
z
bead
F
D
x
DNA
x
bead
i
;
(3.39)
where
z
trap
is the height at the center of the trap, and x
DNA
is determined during
the experiment. Using these data, it was found that the persistence length of DNA,
which quantifies its stiffness, depends on the buffer solution, but is higher than
40 nm, whereas its elastic modulus is of about 1,100 pN.
In addition, the unzipping force of dsDNA was measured with optical tweezers
and found to decrease from about 16 pN to around 10 pN as the number of opened
base pairs increases (
Bockelmann et al. 2002
). Other biomolecules like proteins and
RNA are characterized using this method.
References
Adamcik J, Jung J-M, Flakowski J, de los Rios P, Dietler G, Mezzenga R (2010) Understanding
amyloid aggregation by statistical analysis of atomic force microscope. Nat Nanotechnol
5:423-427
Bhushan B (ed) (2004) Springer handbook of nanotechnology. Springer, Berlin, pp 330-331
























Search WWH ::

Custom Search