Biomedical Engineering Reference
In-Depth Information
A state-of-the-art compromise is achieved by the SHIFTX2 server, which is
able to switch between sequence-based and structure-based methods according
to local prediction quality criteria,
111
yielding RMS errors for
1
H
a
,
1
H
N
,
13
C9,
13
C
a
,
13
C
b
and
15
N shifts of 0.12, 0.17, 0.53, 0.44, 0.52 and 1.12 ppm,
respectively. Although the fidelity of these results is impressive, back-
prediction of chemical shifts is still not sufficiently accurate for highly
stringent applications, such as facile resonance assignment of [
1
H,
15
N]-HSQC
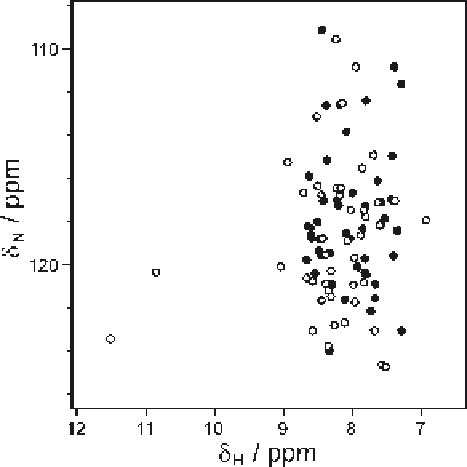
spectra for proteins of known structure. Figure 3.7 displays experimentaland
SHIFTX2 predicted chemical shifts for the backbone amide sites of an a-
helical protein; too few signals are reproduced closely enough to permit a
straightforward
transfer
of
assignment
information
from
back-predicted
frequencies.
Although modest improvements are expected as cross-referenced chemical
shift and structure coordinate data accumulates, discrepancies will probably
remain due to limiting factors which include minor deviations from ideal
geometry, the accuracy of atom positions, bond lengths and bond angles in
experimental protein structures, and contributions from backbone and side-
chain dynamics. Protein motions can be taken into account to some extent by
averaging chemical shift predictions over members of an ensemble of NMR
solution structures
119,120
or snapshots from MD simulation trajectories.
121,122
For example, in tests with a fragment of the ankyrin repeat protein IkBa,
SHIFTX predictions from the static X-ray structure returned RMS errors for
Figure 3.7
Scatter plot comparing the experimental (
#
) and SHIFTX2-predicted (
$
)
backbone amide
15
N and
1
H
N
chemical shifts for a dimeric 50 residue a-
helical protein (M. Ali and R.W. Broadhurst, unpublished results).