Biomedical Engineering Reference
In-Depth Information
More recent algorithms that aim to increase the accuracy of the consensus
CSI method include: the PSSI probability-based approach;
78
the psiCSI
combined sequence alignment/chemical shift-based approach;
79
the PECAN
statistical energy function approach;
80
the TALOS+ neural network/fragment
matching approach;
31
and the DANGLE Bayesian fragment matching
approach.
81
The real challenge for such procedures is to improve on the
performance of secondary structure prediction routines that rely solely on
amino acid sequence alignments. In a recent test against a panel of 29
proteins,
81
the sequence-only JPRED3 server
82
returned the same three-state
classification as the secondary structure identification program DSSP
83
for
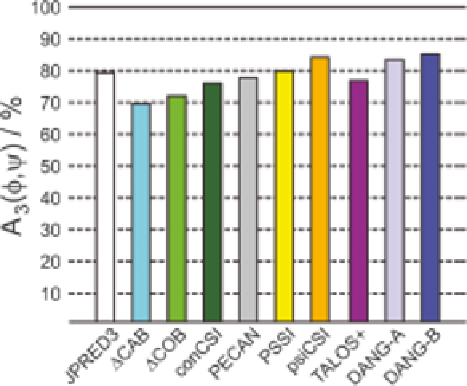
79 % of all residues. As Figure 3.4 shows, taking chemical shift data into
account conferred a clear advantage for only two methods: PsiCSI (84%) and
DANGLE (83-85%). These results fall close to the expected limit for three-
state prediction routines, reflecting the level of discrepancy encountered when
benchmarking different secondary structure detection algorithms or analysing
the variation within an ensemble of solution structures.
77,84
The majority of
disagreements between DANGLE and DSSP occur near protein N- or C-
termini (where the backbone is likely to be flexible), in linker regions that
connect elements of regular secondary structure (where polypeptide chains
often sample multiple conformations), or concern the initiation and termina-
Figure 3.4
Histogram showing A
3
, the percentage of three-state predictions of
secondary structure that agree with the output from DSSP,
83
comparing
various prediction methods. Results are for all residue types in the 29
protein test set described in Cheung et al.
81
Methods are: JPRED3;
82
DCAB;
67
DCOB;
68
CSI;
75
PECAN;
80
PSSI;
78
psiCSI;
79
consensus
TALOS+;
31
DANG-A: DANGLE with all predictions accepted;
81
and
DANG-B: DANGLE after rejecting multi-island predictions.
81