Biomedical Engineering Reference
In-Depth Information
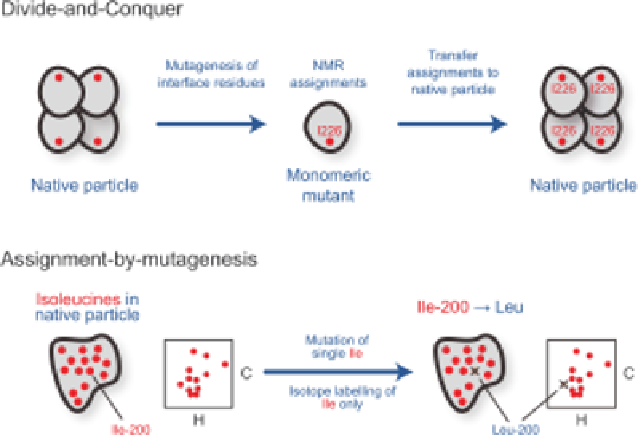
Figure 1.6
Schematic representation of strategies for resonance assignment of methyl
groups in large proteins and protein assemblies. (Top) homo-oligomeric
(or multi-domain) proteins can often be broken into smaller more NMR-
compatible fragments (e.g., a monomeric mutant) that are amenable to
standard backbone and side chain resonance assignment strategies.
Assignments of the smallest species are then transferred back to the native
size protein or complex. (Bottom) an alternative approach centres on the
introduction of mutations into the full-size assembly. For example, in a
[d
1
-
13
CH
3
]isoleucine-labelled protein, a single site-specific Ile
A
Leu
mutation would cause a single resonance to disappear from a 2D
(
1
H,
13
C) methyl HMQC spectrum. Overlaying wild-type and mutant
spectra identifies the missing residue (shown with a cross) and thereby
provides an assignment for the mutated residue.
methyl-TROSY spectra could be acquired when the a-subunits were [U-
2
H],
Ile-[d
1
-
13
CH
3
], Leu,Val-[
13
CH
3
,
12
CD
3
]-labelled and the b-subunits unlabelled.
To obtain sequence-specific assignments, however, it was necessary to dissect
the complex into more tractable pieces. Mutations that stabilised the
monomeric a-subunit or the a
7
-ring were identified. Sequence-specific back-
bone and methyl group assignments were obtained from the 20 kDa
monomeric a-subunit and then transferred to the a
7
-ring and finally to the
full 20S a
7
b
7
b
7
a
7
complex. Using these assignments it was possible to map
intermolecular
complex.
45
interfaces
in
the
1
MDa
activator-proteasome
(Figure 1.1).
The dissection of large homo-oligomeric proteins can require a considerable
amount of trial and error to find optimal mutants or conditions that
sufficiently destabilised oligomeric interfaces without significantly disrupting
the structure of the monomeric building block.