Biomedical Engineering Reference
In-Depth Information
We used as starting conditions the equilibrium concentrations obtained
from the numerical integration of eqn (10.26):
M
I
(0)~(
½
I
eq
,0)
M
IS
(0)~(
½
IS
eq
,0)
M
IC
(0)~(
½
IC
eq
,0)
ð
10
:
30
Þ
Eqn (10.29) was solved for different equilibrium conditions to obtain the
FIDS for the individual species. The FIDS were co-added, zero-filled, and
transformed using a complex Fourier transform to obtain the spectra as shown
in Figure 10.8.
The results show the fast-exchange shifts, but also some small exchange line-
broadening effects of the order of 1 Hz (the linewidths for the isolated species
were taken as 1 Hz).
Where would the auxiliary, independent site S, from which IP6 can easily
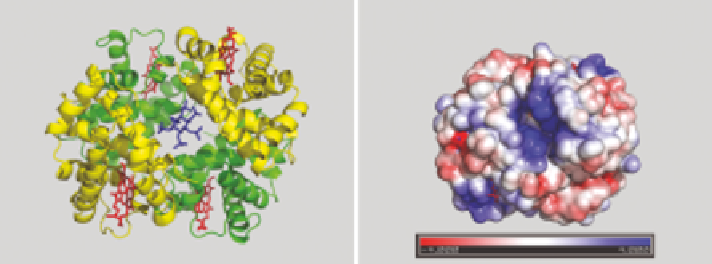
migrate to the cavity site be located? Figure 10.9 shows the structure of deoxyHb
with the location of the IP6 cavity binding site 12. The Poisson-Boltzmann
surface charge calculation, using the facility in Pymol,
17
shows a strongly
positively charged cavity, and this area appears large enough to accommodate
an extra molecule of IP6 on its edge even when the cavity site is filled.
Are the obtained (needed) parameters physically realistic? For the equilibrium
parameters K
D1
5 1.0 6 10
23
M, K
D2
5 1.0 6 10
26
M and K
D3
5 1.0 6 10
29
M, with [Hb]
tot
5 10
23
M, the computations required the kinetic parameters k
1f
52.0 6 10
10
M
21
s
21
, k
1b
5 2.0 6 10
7
s
21
, k
2f
5 1.0 6 10
13
M
21
s
21
, k
2b
5 1.0
6 10
7
M
21
s
21
, k
3f
5 1.0 6 10
9
M
21
s
21
and k
3b
5 1.0 s
21
. k
1f
, which
represents the binding of IP6 to the auxiliary S site is 2.0 6 10
10
M
21
s
21
, right at
the theoretical maximum for diffusion control. This is not unreasonable, since
the hyper-negatively charged IP6 (28 at neutral pH, see previous section)
Figure 10.9
(a) IP6 (blue) modelled in the central cavity of deoxy hemoglobin
(2DN2.pdb) (only a structure for IP6 bound to Hb(CO)
4
is currently
available). The a-chains are green, the b-chains are yellow, and the
hemes are in red. (b) A Poisson-Boltzmann surface charge calculation
for deoxyHb (2DN2.pdb), using the plug-in available in Pymol.
17