Biomedical Engineering Reference
In-Depth Information
13
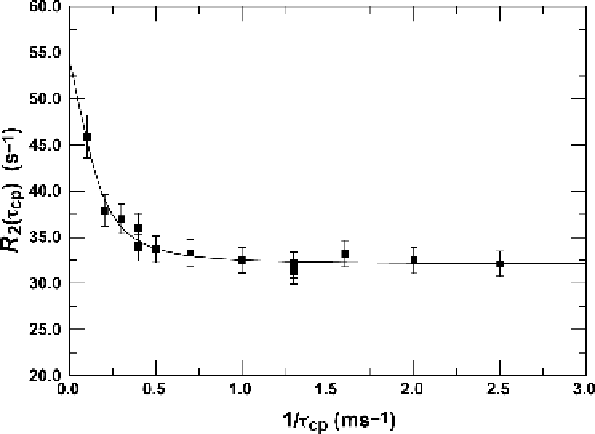
C MQ-CPMG relaxation dispersion for I83d in IGPS bound to
PRFAR. The experiment was acquired at 14.1 T and 303 K. Dispersion
data was fit with eqn (7.11) and gives k
ex
5 500 ¡ 220 s
21
.
Figure 7.4
dispersion data with eqn (7.11) gives a k
ex
5 500 ¡ 220 s
21
, which is similar to
that for the amide backbone (Figure 7.3). The similarity in exchange rate
constants between side-chain and backbone positions further suggests a
concerted motion for these residues and also provides a more complete picture
of ms motions than the individual experiments.
7.5.2 Triosephosphate Isomerase
Triosephosphate isomerase (TIM) is a 53 kDa homodimeric enzyme of the
glycolytic pathway. TIM catalyzes the reversible isomerisation of glyceralde-
hyde-3-phosphate (GAP) and dihydroxyacetone phosphate (DHAP). TIM has
served as a model for understanding the role of conformational changes in
enzyme function. TIM possesses a highly conserved 11 amino acid active-site
loop (loop 6) that must remain open to bind substrate and release product but
that also must close and sequester the active site during the chemical
reaction.
68-72
This structural information coupled with biochemical study of
the catalytic rate for this reaction places a lower limit on the timescale for this
loop movement; the loop cannot move slower than the overall catalytic
turnover rate. This timescale, unfortunately places loop 6 motion outside the
regime to which the CPMG dispersion experiment is sensitive. Its motion was
therefore characterised with the TROSY-selected off-resonance R
1r
experi-
ment (Figure 7.5).
7,73
Fits of the R
1r
dispersion data for the N-terminal loop 6
residue V167, with the fast-limit form of eqn (7.8), provided a k
ex
value equal
to 8900 ¡ 1600 s
21
, a value that is nearly identical to k
cat
for this enzyme. This