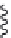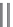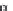Biomedical Engineering Reference
In-Depth Information
O
OH
H
H
2
N
O
Valine
Amide
O
OH
O
HO
O
-Hydroxy-valine (Vah)
Ester
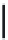
FIGURE 4.9
By using conventional genetic methods, changes in the protein backbone is not possible;
however, by applying either unnatural mutagenesis or protein ligation strategies the amide backbone can be
changed into, for example, an ester by using a-hydroxy acids rather than amino acids. This can be used to
evaluate electronic effects of backbone carbonyl groups.
sensitive detection systems, such as electrophysiology or l uorescence, are required in these studies.
Thirdly, the technology is generally limited to
in vitro
systems. Thus, in order to overcome some
of these limitations, a modii ed method for incorporation of unnatural amino acids into proteins
in vivo
, was introduced. In this approach a custom-made pair of tRNA and aaRS is genetically
introduced into a cell and the aaRS is engineered so that it only recognizes the unnatural amino acid
and efi ciently acylates the corresponding tRNA. Subsequently, the unnatural amino acid, which
has to be nontoxic and cell permeable, is added to the growth media, taken up by the host organism
and incorporated into the protein by the specii c tRNA/aaRS pair. This technology, has been suc-
cessfully applied in both yeast and eukaryotic cell, and allows the generation of proteins with an
unnatural amino acid in reasonable yields. The primary challenge of this technology is that specii c
aaRS have to be generated for each unnatural amino acid, which is done by extensive mutational
studies and rounds of positive and negative selections.
The technology has been applied to a number of model proteins, and has been used to specii -
cally incorporate a glycosylated amino acid into myoglobin. In addition, a fully autonomous bac-
terium,
E. coli
, has been engineered so it could synthesize
p
-amino-phenylalanine, and a specii c
tRNA/aaRS pair was introduced, which allowed incorporation into myoglobin. The technology also
holds commercial prospective, and a company (Ambrx) is developing protein therapeutics based on
this technology.
A general limitation of these technologies is that the genetic code only contains three stop
codons, which limits the theoretic numbers of different unnatural amino acids, that can be incor-
porated in a single protein to two. To overcome this limitation, Sisido and colleagues have explored
an alternative strategy using extended codons and frameshift suppression. In this approach, an
mRNA containing an extended codon consisting of four or i ve bases is being read by a modii ed
aa-tRNA containing the corresponding extended anticodon. In certain species, some naturally
occurring codons are rarely used and the amount of their corresponding tRNA is low. This has
been used in the design of four-base codons, which are derived from these rarely used codons,
to minimize the competition between the four-base anticodon tRNA and endogenous tRNA. The
four-base codon technique has been used to incorporate unnatural amino acids into proteins in
E. coli
. It has also been used to incorporate two different unnatural amino acids into two differ-
ent sites of a single protein showing that four-base codons are not only orthogonal to their host
organism but also to each other.