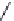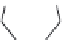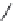Biomedical Engineering Reference
In-Depth Information
“Structural water”
Ph
MeO
OH
N
HO
OH
H
OH
Ph
HO
OMe
H-bond donor/acceptor
(A)
(B)
“Structural water”
O
O
O
R
NN
R
P'
P'
P
1
P
1
P'
P
1
OH
HO
OH
HO
OH
H-bond donor/acceptor
(C)
(D)
(E)
FIGURE 2.9
The DuPont Merck design process leading to the seven-membered urea HIV-1 protease inhibi-
tors. (A) Symmetric dihydroxyethylene inhibitor used to dei ne a pharmacophore. (B) Initial hit from 3D data-
base search. (C) Initial synthetic scaffold. (D) Scaffold modii ed to accommodate two hydrogen-bond donors/
acceptors. (E) Final scaffold optimized for synthetic feasibility and improved hydrogen bonding.
designing an inhibitor that would displace this water molecule, a favorable entropy contribution to
the binding energy would be obtained. In addition, selectivity should be gained since this structural
water molecule was unique for the viral proteases.
The DuPont Merck scientists dei ned a pharmacophore (see Chapter 3) from known dihydroxy-
ethylene inhibitors (Figure 2.9A), used this pharmacophore for searching a database and obtained
a hit (Figure 2.9B), which subsequently gave the idea to the six-membered cyclic ketone as lead
structure (Figure 2.9C). Ring expansion to the seven-membered cyclic ketone (Figure 2.9D) enabled
incorporation of two hydrogen-bonding donor/acceptor groups and synthetic reasons led to the
HN
NH
IIe51'
IIe51
O
Np
N
N
Np
Bn
Bn
HO
OH
O
Asp25'
O
O
Asp25'
O
Asp25
Asp25
(A)
(B)
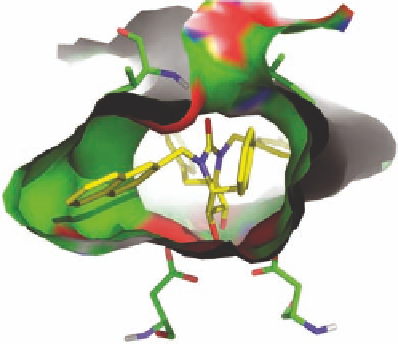
FIGURE 2.10
Schematic representations of the binding of the seven-membered urea HIV-1 protease inhibi-
tor XK-263 to HIV-1 protease (pdb-code 1HVR). In (A), Bn and Np refer to benzyl and 2-naphthyl, respec-
tively. In (B), the two Asp25 and Ile51 residues making hydrogen bonds to the inhibitor are shown as stick
models. Heteroatoms are colored red and blue for oxygen and nitrogen, respectively. The surface is colored
accordingly.