Biomedical Engineering Reference
In-Depth Information
of each the following histones; H2A, H2B, H3, and H4. The basic repeating unit of eukaryotic DNA
is termed the nucleosome, and consists of 146 bp of DNA wrapped 1.65 left-handed turns around a
core histone particle. The N-terminal tails of the core histones protrude away from the core histone
particle as schematically presented in Figure 23.6A. These tails, especially those of histone H3 and
H4, are subjected to a number of posttranslational modii cations (PTMs), including arginine and
lysine methylation, serine phosphorylation, and lysine acetylation amongst others.
The pattern of histone PTMs together with the pattern of DNA methylation is termed the
epigenetic makeup or epigenetic code of the cell. One important form of histone PTM is lysine
ε
-
N
-acetylation, which is a highly dynamic process, and the result of the opposite activities of his-
tone acetyl transferases (HATs) and histone deacetyl transferases HDACs (Figure 23.6A). HDAC
Ac
+
+
+
+
NH
3
NH
3
Ac
NH
3
HDAC
+
NH
3
NH
3
+
NH
3
Ac
Ac
Ac
+
+
Ac
Ac
+
NH
3
NH
3
Ac
Ac
Ac
NH
3
+
+
+
NH
3
NH
3
NH
3
NH
3
Ac
Ac
+
+
HAT
Ac
NH
3
NH
3
Ac
Ac
Ac
(A)
O
O
H
NH
S
HN
S
O
O
O
OH
O
H
NH
O
O
Depsipeptide (FK228)
Tricostatin A (TSA)
O
O
O
-
Na
+
N
OH
H
O
SAHA, Vorinostat
Sodium valporate
O
O
NH
2
H
N
N
N
S
OH
N
O
O
O
O
(B)
MS275
PXD101, Belinostat
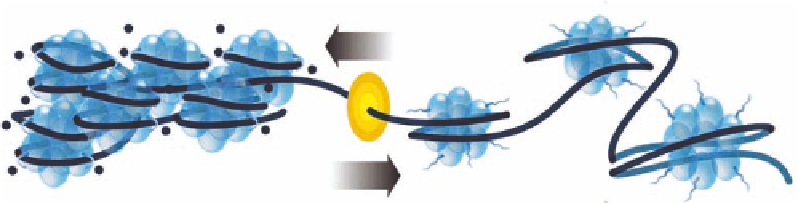
FIGURE 23.6
(A) Schematic representation of core histone complexes on DNA, showing the effect of opposing
HAT and HDAC activities on the acetylation status of the
N
-terminal tails of core histones. (B) Chemical struc-
tures of some HDAC inhibitors in clinical development.