Biomedical Engineering Reference
In-Depth Information
Lowest energy conformation in aqueous solution
Bioactive conformation
+10.5 kJ/mol
(a)
(b)
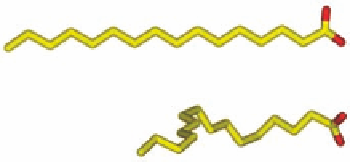
FIGURE 1.4
(a) Palmitic acid bound to the adipocyte lipid-binding protein (pdb-code 1LIE) and (b) the
preferred conformation of palmitic acid in aqueous solution and the conformation bound to the protein.
apo-form
apo-form
C-loop
C-loop
(a)
(b)
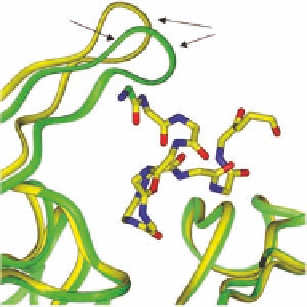
FIGURE 1.5
(a) Epibatidine (orange) bound to the AChBP. The orange C-loop belongs to the epibatidine-
AChBP complex (pdb-code 2BYQ), the green C-loop belongs to the uncomplexed AChBP (apo-form, pdb-
code 2BYN). (b) α-conotoxin ImI (yellow carbons) bound to AChBP (pdb-code 2BYP). The yellow C-loop
belongs to the α-conotoxin ImI-AChBP complex, the green C-loop belongs to the uncomplexed AChBP.
note that in calculations of conformational energy penalties, the conformational properties of the
unbound ligand “in aqueous phase” must be used as the reference state (see Further Readings).
In terms of
G
conf
, rigid molecules have an advantage relative to more l exible ligands. The
binding of a rigid ligand does not result in a loss of conformational entropy and if the ligand
has only one possible conformation (or has one strongly preferred conformation correspond-
ing to the binding conformation), the conformational energy penalty is zero. Although highly
rigid molecules are ideal as ligands, it is a great challenge to design such ligands. For instance,
functional groups taking part in interactions in the binding cavity have to be designed to occupy
precisely correct positions in space, as no (or very small) adjustments of their positions are pos-
sible in a rigid ligand.
So far only conformational changes in the ligand have been discussed, but conformational
changes most often also occur in the protein. Not only may the amino acid side chains adjust their
conformations to optimize their interactions with the ligand, but the protein backbone conforma-
tion may also change. In some cases, this may result in major movements of, for example, loops
or even entire protein domains. Examples of such movements are shown in Figure 1.5. The C-loop
in the acetylcholine binding protein (AChBP) and most probably also the corresponding loop in,
Δ