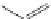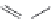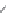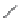Biomedical Engineering Reference
In-Depth Information
N
N
N
+
N
O
O
OH
O
O
O
O
O
OH
O
O
OH
O
OH
Atropine (
16.37
)
Scopolamine (
16.38
)
NMS (
16.39
)
QNB (
16.40
)
O
H
N
N
H
N
H
O
Methoctramine (
16.41
)
O
CN
N
O
N
O
N
N
N
N
N
N
S
O
O
SCH 57790 (
16.44
)
O
N
N
N
N
N
Pirenzepine (
16.42
)
AF-DX-116 (
16.43
)
N
C
O
N
S
C
*
O
O
(
16.46
)
m2-toxin (
16.45
)
FIGURE 16.9
Chemical structures of competitive mAChR antagonists. (Part of the i gure is reprinted from
Krajewski, J.L. et al.,
Mol. Pharmacol
., 60, 725, 2001. With permission.)
16.4.3 A
LLOSTERIC
M
ODULATORS
OF
mAChRs
The difi culties connected with the development of agonists or competitive antagonists capable
of discriminating between mAChR subtypes have prompted the search for and identii cation of
“allosteric modulators” of the receptors (Chapter 12). As can be seen from the examples given in
Figure 16.10, these compounds (
16.47-16.51
) are structurally completely different from ACh and
other orthosteric mAChR ligands in a complete manner. Interestingly, several of these allosteric
modulators also target other receptors or enzymes.
16.4.4 L
IGAND
B
INDING
TO
THE
mAChR
A considerable insight into the ligand-binding modes of mAChR ligands to their receptors has, over
the years, been gained from mutagenesis studies. The orthosteric site in the mAChR (and other fam-
ily A GPCRs) is localized in a cavity formed by transmembrane regions (TMs) 3, 5, 6, and 7 (Figure
16.11A). All orthosteric mAChR ligands contain a quaternifed ammonium group or an amino group