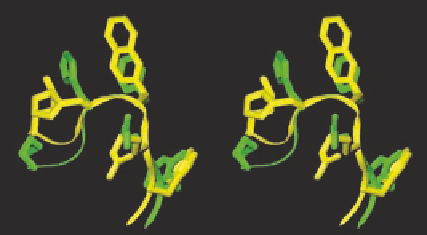Biomedical Engineering Reference
In-Depth Information
Peptide linear presentation of
pharmacophore
Peptide 3D presentation of
pharmacophore
Peptide turn presentation of
pharmacophore
FIGURE 8.8
Peptide structures to mimic in peptide mimetic design.
FIGURE 8.9
NMR structures of superimposed MTII (green) and SHU9119 (yellow) (left) and NMR struc-
ture of AGRP (right).
peptides with powerful biological properties. The use of computational methods in conjunction with
molecular modeling is a powerful tool in design considerations, with the caveat that for novel struc-
tures, current force i elds might not give an accurate picture of the structure or energetics of the
system. As an example of melanocortin system, along with the nuclear magnetic resonance (NMR)
structure of Agouti-related protein (AGRP, an endogenous antagonist of hMC3R and hMC4R) and
MTII (superagonist of hMC4R), the 3D pharmacophore of human melanotropin has been partially
deciphered (Figure 8.9). The primary structure of both MTII and AGRP are quite different, but
from their NMR it can be found that they both share the same
-turn like structure. These accom-
plishments combined with the computational chemistry can be very crucial for designing novel
selective agonist and antagonist compounds.
There is one computational approach for mapping molecular interaction space without knowl-
edge of the 3D structure, known as proteochemometrics. It is an extension of traditional ligand-
based 3D QSAR approaches. It exploits afi nity data for a series of diverse organic ligands binding
to different receptor subtypes, providing proteochemometric analysis, and insights into ligand
recognition.
β
8.3.2 D
ESIGN
B
ASED
ON
THE
B
IOLOGICAL
F
UNCTION
The paradigm that underlies the chemogenomics approaches to ligand design can be stated as
“similar receptor/acceptor binds similar ligand.” This implies that for a novel receptor/acceptor, the