Biomedical Engineering Reference
In-Depth Information
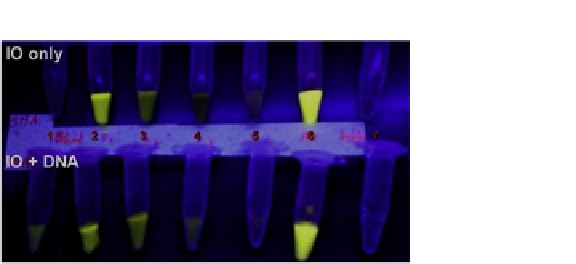
FIGURE 7.2
Amine functionalized IOMNP were used to capture dsDNA that was from lysed
SK-BR3 cells. (Legend: 1-IO after dsDNA extraction, 2-supernatant 1 (SUP1) after capture and
acridine orange (AO) addition, 3-SUP2 after capture+AO, 4-SUP3 after capture+AO, 5- SUP4 after
capture+AO, 6-AO +buffer, 7-buffer).
Courtesy of Ocean NanoTech. (For color version of this
figure, the reader is referred to the online version of this topic)
7.3.3.1 Protocol for the Electrostatic Loading of dsDNA in IOMNPs
The following protocol is used to prepare and verify the formation of
dsDNA~IOMNP complex. The dsDNA was prepared from SK-BR3 breast can-
cer cell lines.
(1)
Grow the SK-BR3 cells following manufacturer's protocol.
(2)
When the cells reach 70-80% confluence, harvest following the manufac-
turer's protocol.
(3)
Place 100,000 cells in a microcentrifuge tube.
(4)
Centrifuge at 4000 rpm to precipitate the cells.
(5)
Remove the supernatant and add 100 uL of Dulbecco's Phosphate-
Buffered Saline (DPBS).
(6)
Lyze the cells with lysozyme or sonicate for 15 min to rupture the cells.
(7)
Take 0.5 mg of 30 nm IOMNPs that are amine functionalized (Ocean
NanoTech Catalog # SHA30).
(8)
Magnetize to isolate the IOMNPs from solution.
(9)
Disperse in 100 uL deionized water (DI) water and adjust the pH to 5.
(10)
Add the lyzed cells into the IOMNPs and vortex.
(11)
Incubate in a slow shaker for 1 h.
(12)
Centrifuge at 5000 rpm for 10 min.
(13)
Discard the supernatant and save the pellet which consists of the
IOMNP~dsDNA.
(14)
Wash the IOMNP~dDNA three times with sodium citrate buffer (1×SSC).
(15)
Reconstitute with 150 uL DI water.
(16
) Add 50 uL of acridine orange.
(17)
Put in a shaker for 20-30 min.
(18)
Centrifuge at 5000 rpm for 10 min.
(19)
Discard the supernatant and save the pellet which consists of the
IOMNP~dsDNA+acridine orange.