Information Technology Reference
In-Depth Information
Fig. 2.
An actin-agent can be in three different states. A free molecule can bind to an already
bound initial agent (
BB = 0
). The already bound actin-agent then become fully bound. Then the
third actin-agent can bind to the second actin-agent which become fully bound and so on.
molecule binds to another, then the identification number of the counterpart is stored
in the BB (respectively TB) variable; the agent becomes immobilised and its rotation
will be adapted. The precondition for binding is, that one of the agents is already bound
(bottom-bound). This leads to the condition that at least one agent has to be stuck in
the beginning of the simulation. This is done by initialising one agent with
BB = 0
.
If a free agent binds to an already bound one, the second becomes then fully bound
(
BB
=
−
1
,
TB
=
−
1
). The whole schema of the actin-actin interactions is also shown
in Figure 2.
The polymerisation of actin filaments is characterised by a 70
◦
angle branching on
several positions mediated by the Arp2/3 protein [25]. To simulate this branching pro-
cess, a new agent with a third binding side was implemented. The orientation of the
branching side to the left or right was set randomly. This agent is restricted to bind
only to actin-agents, so that a Arp2/3-Arp2/3 combination is prohibited. In Figure 3
the scheme for the interactions and state changes is illustrated. Similar to the actin-
agent, the size of this agent was determined from published measurements [24]. Figure
4 shows the approximated dimensions of Arp2/3.
An agent-based model has to include the reaction kinetic in a reasonable way. Due
to the nature of spatial simulations with individual molecules, this may be done by
an interaction volume, which defines a reaction zone around a particular agent (see
Figure 5). Andrews and Bray (2004) developed an algorithm to determine this volume,
but considered more detailed interactions. Another way is described by Pogson et al.
(2006) where the interaction radius
r
is calculated by:
r
=
3
3k
Δt
4
π
N
A
10
3
where k is the kinetic rate constant,
Δt
the discrete time interval and
N
A
is Avogadro's
constant (
6
.
022
×
10
23
). The rate constant for actin-actin assembly was determined with
11
.
6
μM
−
1
s
−
1
[7] and leads to a radius of 0.166
˚
Afor
Δt
=1
s
. If two or more agents
enter the interaction volume at the same time step, the closest molecule to the reaction
molecule assembles to it, if two or more have the same distance, one will be chosen by
chance.


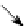























Search WWH ::

Custom Search