Biology Reference
In-Depth Information
field has already been used to simulate important challenges in membrane simula-
tions such as estimating timescales for concerted lipid motions (
Apajalahti et al.,
2010
), characterizing lipid rafts from a molecular perspective (
Risselada &
Marrink, 2008
) or studying preferential interaction between lipid species
(
Marrink, de Vries, Harroun, Katsaras, & Wassall, 2008
). In addition, along with
pure membrane simulations, various studies have used the MARTINI force field
to characterize the tendency of certain TM proteins to partition into lipid domains
of different composition (
Sch¨fer et al., 2011
). Interestingly,
Doma
nski, Marrink,
and Sch¨fer (2012)
have recently simulated the lateral heterogeneity of biological
membranes by using the MARTINI force field and showed how the protein bacte-
riorhodopsin partitions into liquid-disordered lipid domains. This study is a perfect
example of how CG simulations can be used to simulate GPCRs in its native-like
environment prior to study receptor-receptor interactions.
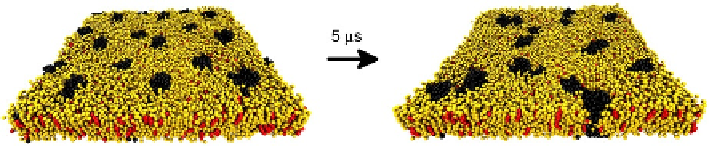
CG simulations enable to simulate a relatively large set of GPCRs embedded in a
membrane patch (
Fig. 4.4
). In these simulations, GPCRs can freely diffuse across the
membrane and transiently or stably interact with other GPCRs, that is, we can sim-
ulate the self-aggregation of GPCRs (
Periole et al., 2007
) and, what is more, we can
use more native-like membranes when modeling such self-aggregation. As for all-
atom simulations, CG simulations of GPCRs benefit from the accuracy of both an
adequate protein input structure and a sufficiently equilibrated lipid environment.
Thus, an ideal protein input structure should be based on the information contained
in any of the currently solved GPCR crystal structures. One of the interesting tools
made available by the MARTINI force field team (
http://md.chem.rug.nl/cgmartini/
)
FIGURE 4.4
Coarse-grained (CG) simulations of GPCRs embedded in membranes. This figure represents
two snapshots of a CG simulation of 18 GPCR monomers embedded in a membrane.
This system was created by multiplying a unit cell consisting of two GPCR monomers
embedded in a smaller membrane patch. Membrane lipids were first equilibrated around
proteins by restraining all GPCR monomers and running the system during 100 ns. The final
snapshot of this equilibration run (left picture) is the starting configuration of the production
run. GPCR monomers can now freely diffuse and rotate to find each other and ultimately
self-assemble (right picture). Simulation runs longer than 5 ms (i.e., above 10 ms) are normally
needed to consistently study GPCR oligomerization. Water and ions are not displayed for
clarity; GPCR monomers are depicted in black, whereas phospholipids and cholesterol
are depicted in yellow and red, respectively.