Biology Reference
In-Depth Information
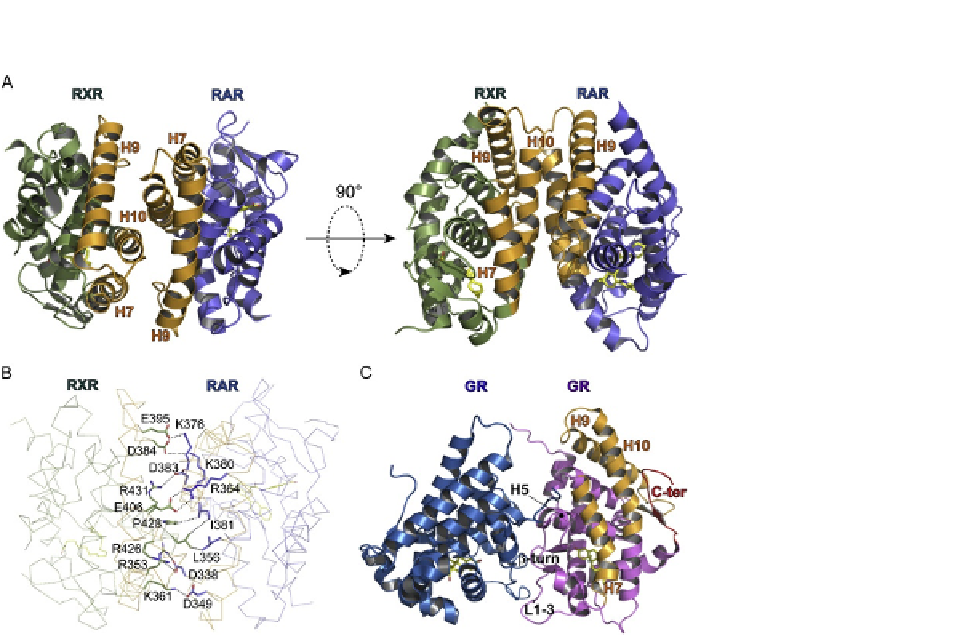
FIGURE 2.2
Structural analysis of NR dimers. (A) The dimeric arrangement of NRs is illustrated by the
structure of RARa-RXRa LBD heterodimer (Protein Data Bank code 1dkf) viewed along
(left) or perpendicular (right) to the dimer axis. The secondary structural elements involved in
the dimer interface are displayed in orange and labeled. The ligands in both subunits are
represented as yellow sticks. (B) Some important intersubunit interactions of the
RARa-RXRa LBD heterodimer are shown. For clarity, not all the contacts are displayed.
(C) The dimeric arrangement of the GR LBD homodimer as seen in the crystal (Protein
Data Bank code 1m2z). The secondary structural elements involved in the canonical
dimerization surface used by other NRs are labeled together with the C-terminal b-strand that
masks a portion of this surface. The structural elements that are involved in the GR-GR
interaction are indicated.
based on crystal structure and sequence alignment, has been reported (
Bourguet, Vivat,
et al., 2000
). The reader is referred to this publication for details and an interpretation
of the structural basis that accounts for the homo- and heterodimerization pattern of
distinct members of this family. Briefly, the structures show that the dimeric arrange-
ments are closely related, with residues from helices H7, H9, and H10 and loops L8-9
and L9-10 of each protomer forming an interface comprising a network of comple-
mentary hydrophobic and charged residues and further stabilized by neutralized basic
and acidic surfaces (
Fig. 2.2
Aand
2.2
B). Interestingly, the recently reported crystal
structures of the entire PPAR
g
-RXR
a
heterodimer and HNF-4
a
homodimer bound