Biology Reference
In-Depth Information
constant for the image acquisitions of all samples in a given experiment (experiment
and monomeric control).
Note that when applying SpIDA to fixed samples for single-image analysis, it is
better to increase the laser intensity, always considering the effect of photobleaching,
than the PMT gain/voltage because raising the PMT gain will result in increased de-
tector variance.
In all cases, image acquisitions can be single image, time series, or Z-stack.
SpIDA may be applied to measure proteins expressed at the plasma membrane or
in other cellular organelles, with the caveat that the spatial resolution is set by the
three-dimensional (3D) PSF of the CLSM.
1.3.4
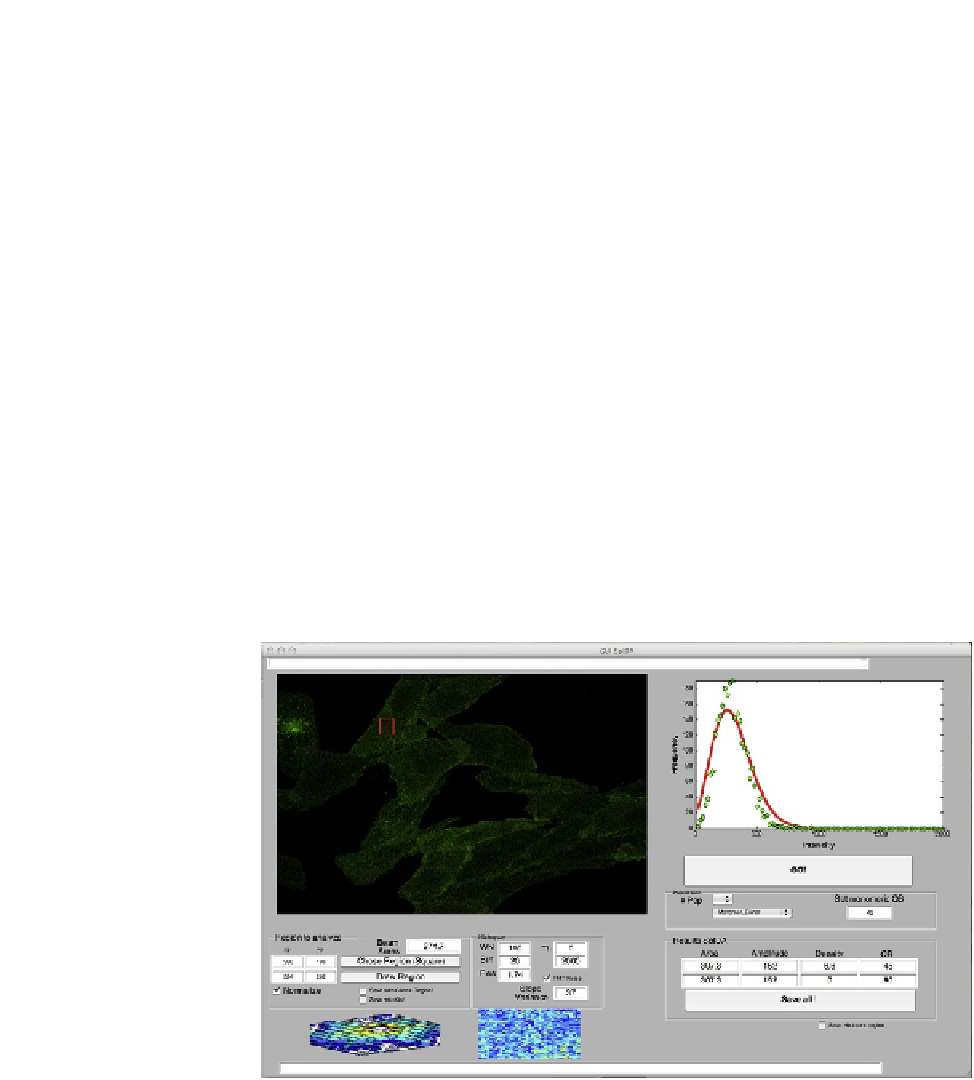
Analysis with the SpIDA program graphical user interface
A customMATLAB graphical user interface (GUI) was designed to perform SpIDA
in a user-friendly way (
Fig. 1.7
). This section provides basic information for oper-
ating the GUI.
The GUI can be downloaded free of charge at the website
http://www.
neurophotonics.ca/en/tools/software
.
The GUI was tested in the Windows 32- and
64-bit MAC OS X, and UNIX operating systems and was created in MATLAB
FIGURE 1.7
MATLAB GUI for SpIDA.