Agriculture Reference
In-Depth Information
early generations of crosses have been proposed by Jinks
and Pooni.
They have shown that for any continuously varying
character, the expected mean and variance of all possible
inbred lines, derived by inbreeding following an initial
cross between two homozygous parents, can be specified
in terms of the components of means and variances as
specified by biometrical genetics. For example, if an
additive-dominant genetic model of inheritance proves
adequate, the expected mean is
m
, the mid-parent value,
and the expected variance of the inbred sample is
V
A
.
From the predicted mean and variance we can deter-
mine many of the properties of the recombinant inbred
lines that can be derived in a pure-line breeding pro-
gramme based on the performance of generations in
the early generation stages. In addition, the relative
probabilities with which different pair-wise crosses will
produce inbred lines with particular properties can also
be predicted and hence used as a selection criteria
for reducing the number of breeding lines in a plant
breeding programme.
The crosses which show highest probability of pro-
ducing desirable recombinants can therefore be iden-
tified from those with a lesser chance of producing
desirable lines. Rather than selecting individual geno-
types at the early generation stage, the number of
surviving lines can be reduced by selection of the supe-
rior cross combinations. Similarly, if the probability of a
desirable recombinant is known from a particular cross,
then this value can be used to determine the number
of recombinants which need to be evaluated to ensure
that 'at least one' is found. When a single trait is exam-
ined, this procedure of estimating and using genetic
parameters is called
univariate cross prediction
.
Univariate cross prediction has been applied to a
number of inbreeding species based on the initial work
of Jinks and Pooni with
Nicotiana rustica
. Predictions
of the proportion of recombinant inbred lines that will
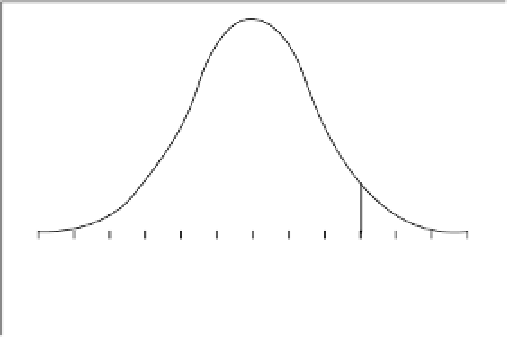
transgress a predefined target value (
T
) are based on the
evaluation of the integral:
∞
∞
f
(
)
∫
,
f
(
,
) d
, d
T
T
Figure 7.12
Illustration of Univariate cross prediction
technique.
Estimation of
m
and
V
A
The additive genetic components of the expected
variance (
V
A
) can be estimated from a number of
different sources initiated by a cross between two
pure-breeding lines. Methods that have proven reliable
include:
•
Producing a sample of inbred lines from a pair-wise
cross between two parents by rapid cycle single seed
descent or doubled haploid techniques
•
The standard P
1
,P
2
,F
1
,F
2
,B
1
and B
2
families
•
A triple test cross, where a random sample of the F
2
from each parent cross-combination is backcrossed to
P
1
,P
2
and the heterozygous F
1
(The theory of the
triple test cross is beyond the scope of this topic.)
•
Evaluation of a random sample of F
3
families
If a sample of inbred lines from a number of different
crosses are grown in properly designed assessment trials,
it is possible to estimate
m
, the average performance
of all possible inbred lines from each cross and
A
, the
additive genetic variation for each cross. The average
performances of the inbreds are a direct estimate of
m
,
and the variance between inbreds (
σ
g
) in the sample
is a direct estimate of
A
after error variance has been
removed.
It is possible to calculate the proportion of lines
expected to transgress a predefined target value (
T
)
by using:
f
(
x
i
)
d
x
i
T
−
m
m
−
T
T
or
σ
g
depending on whether the predictions are for values
greater than (or equal to), or less than (or equal to) the
target value set. Where
T
is the target value,
m
is the
σ
g
where the variate of interest is normally distributed
and the function
f
(
x
i
)
is based on
m
, the mean of
all possible inbreds for a character and
A
, the additive
genetic variance for the character (Figure 7.12).