Biomedical Engineering Reference
In-Depth Information
Figure 20.8
MapReduce
respect by allowing the frameworks to be accessed publicly, for a fee, via
Amazon Web Services. Now the landscape is ripe with tools and
technologies targeted at the NoSQL and Big Data paradigm. Table 20.1
summarizes the database technologies discussed here, and some of their
common characteristics.
Each of the four NoSQL databases store data using a global primary
key/value pair. Each key is scoped to a namespace (also called a bucket or
a partition), which allows the system to identify each piece of data
uniquely. This is logically analogous to the 'CDA Document ID' or the
'CDA Data Item ID' in the 'CDA Data Items' table, in Figure 20.7.
However, the database's intrinsic understanding of this allows each of
these engines to distribute the data for native sharding across distributed
disks and distributed server nodes. This is similar to implementing table
partitioning across the 'CDA Data Item', although in the case of the
NoSQL clustering capabilities, it is built in, easy to administer, and native
to the basic clustering methodology. This also mandates extremely simple
query interfaces for storing and retrieving groups of data. These interfaces
do not allow the messy unpredictable joins, and thus provides a very
uniform buffering and disk IO usage pattern that allows the databases to
be tuned and engineered for high reads. Finally, many of them provide
lightning fast writes due the simplifi cation of the locking mechanisms by
requiring writes to simply be an append operation while versioning the
entire set of data.



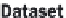



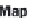


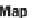




























Search WWH ::

Custom Search