Biomedical Engineering Reference
In-Depth Information
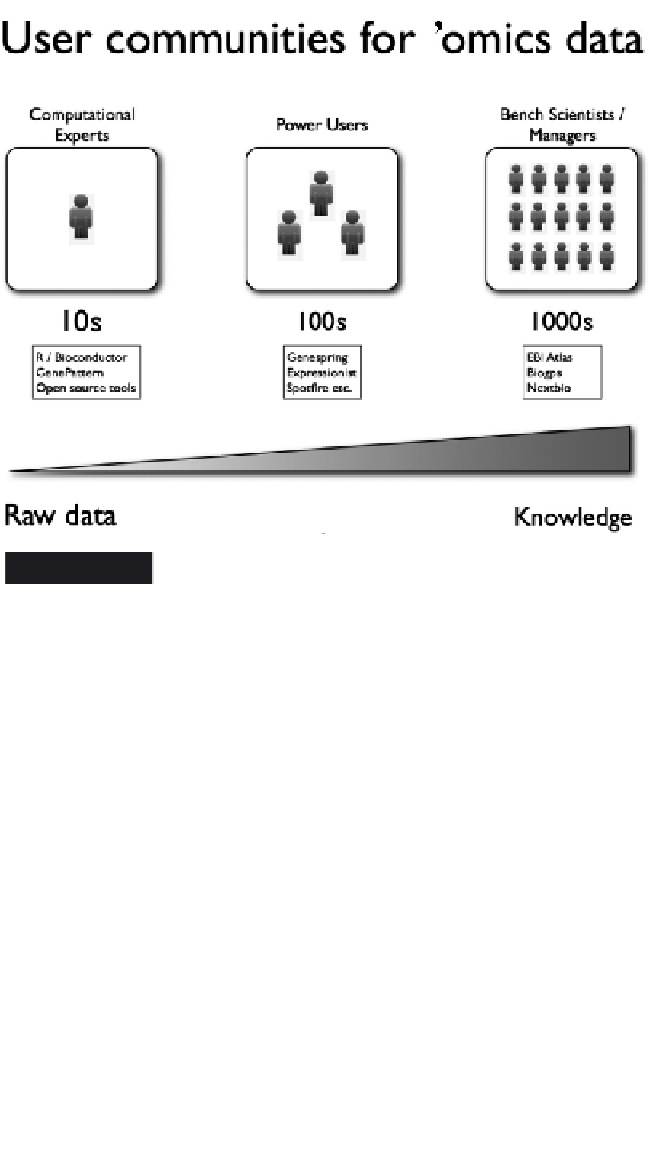
Figure 9.2
User communities for 'omics data
have been undertaken in recent years to bring this situation under
control; establishing the Minimal Information About a Microarray
Experiment (MIAME) [5] and the Minimum Information about a high-
throughput SeQuencing Experiment (MINSEQE) [6] standards for
microarrays and NGS experiments and a common data exchange format,
MAGE-TAB [7].
In order to create a useful 'omics portal, two additional steps are
needed beyond aggregation of standards-compliant data:
1. curation towards a commonality of experimental descriptions across
all available data; and
2. statistical analysis enabling queries over the entire curated data
collection.
Curation is a part-automated, part-manual process, the aim of which is to
prepare data for meaningful downstream analysis, by ensuring data
completeness and correctness and mapping free-text descriptions to
controlled vocabularies, or ontologies. In bioinformatics, ontologies are
used widely to model knowledge of various domains: the OBO Foundry [8]



Search WWH ::

Custom Search