Biomedical Engineering Reference
In-Depth Information
Figure 6.12
Outline of a workfl ow for image processing
analyzed. As this workfl ow is again quite complex, we show the outline
in Figure 6.12.
In the fi rst step, Image Readers are used to load all images from a
directory into KNIME. The Image Processing feature comes with a new
data type for images to enable them to be used in data tables in the same
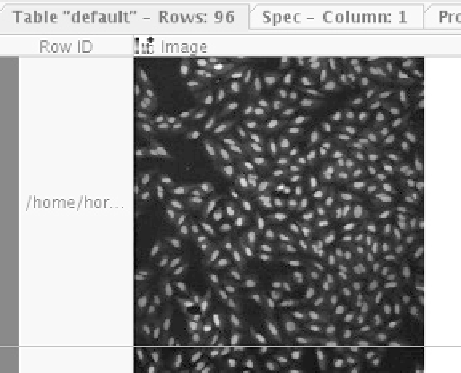
way as strings, numbers, or molecules. Figure 6.13 shows one such image
in KNIME's table view (the images are taken from the public SBS
Bioimage CNT dataset).
As there are two sets of images, one for the nuclei and one for the
cytoplasm, before the features are computed they are combined into one
table. The image features are computed independently for each single
cell. Therefore the cells have to be identifi ed fi rst. This is performed by
taking the nuclei images and applying a binary thresholder to distinguish
the nuclei clearly from the background, see Figure 6.14.
These images are subsequently used as seeds for a so-called Voronoi
segmentation. This process takes the cytoplasm images and segments
them into many different non-overlapping regions around the seeds.
Figure 6.13
Black-and-white images in a KNIME data table




Search WWH ::

Custom Search