Biomedical Engineering Reference
In-Depth Information
does not try to interpret the contents of the fi le but merely extracts the
text representing each molecule into a cell of the output table. The
interpretation is done by special conversion nodes (e.g. CDK, Indigo,
Schrödinger, Tripos, etc.), which are more or less strict about the actual
representation of a molecule. Second, the Concatenate node has so-called
optional input ports. Usually, a node can only be executed if all input
ports are connected, but for optional input ports this is not necessary.
This makes perfect sense when concatenating tables because it frees the
user from having to build a cascade of two-port concatenation nodes
when there are several tables to combine.
The next part of the workfl ow deals with the preparation of the
molecules. The Indigo [4] nodes are used for the whole fi ltering process.
These nodes were recently contributed as open source by GGA Software
and build on their Indigo chemical library.
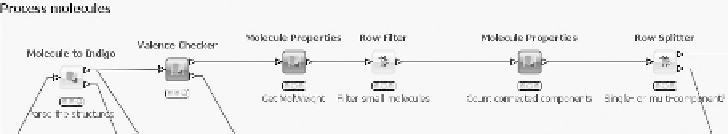
In the fi rst step (Figure 6.6), the SD records read in by the previous step
are converted into the internal Indigo format. Molecules that cannot be
converted, for example because of wrong stereochemistry or invalid atom
types, are transferred to the second output port and added to the list of
problematic structures (not shown). All remaining molecules are checked
for correct valences and structures further down the pipeline. Structures
failing the test are again added to the problematic structures. In the next
step very small compounds (having a molecular weight of less than 10)
are fi ltered out as uninteresting. Also, all structures that consist of several
fragments are removed. This is necessary because in the next step (see
Figure 6.7) canonical SMILES are generated, which cannot handle
disconnected structures.
The most important step involves the Group By node, which groups all
incoming rows based on the canonical Smiles string. In addition, it adds
two columns to each canonical SMILES: a list of IDs that share the same
SMILES, and the number of rows with that SMILES. The following row
splitter is then used to divide the molecules into unique molecules (fi rst
Figure 6.6
Preparation of the molecules



Search WWH ::

Custom Search