Biomedical Engineering Reference
In-Depth Information
to do is download [1] and unpack an archive (which already includes
Java).
With KNIME, the user can model workfl ows, consisting of nodes that
process data, which is transported via connections between the nodes. A
fl ow usually starts with a node that reads in data from some data source,
usually text fi les, but databases can also be queried by special nodes.
Imported data are stored in an internal table-based format, where
columns have a certain data type (integer, string, image, molecule, etc.)
and an arbitrary number of rows conforming to the column specifi cations.
These data tables are sent along the connections to other nodes. In a
typical workfl ow, the data will fi rst be pre-processed (handling of
missing values, fi ltering columns or rows, partitioning into training
and test data, etc.) and then predictive models are built with machine
learning algorithms such as decision trees, naive Bayes classifi ers or
support vector machines. A number of view nodes are available to inspect
the results of analysis workfl ows, which display the data or the trained
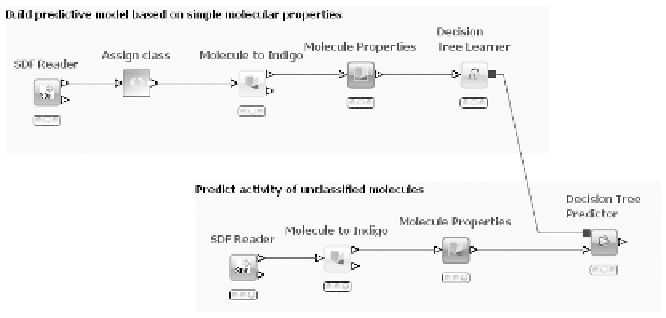
models in various ways. Figure 6.1 shows a small workfl ow with some
nodes.
The fi gure also illustrates how workfl ows can be documented by use
of annotations: in the upper part of the fl ow classifi ed molecules are read
in, properties are calculated, and fi nally a decision tree is built to
distinguish between active and inactive molecules. The lower part reads
unclassifi ed molecules and predicts activity by using the decision tree
model.
Simple KNIME workfl ow building a decision tree for
predicting molecular activity
Figure 6.1



Search WWH ::

Custom Search