Biomedical Engineering Reference
In-Depth Information
method of detecting overfi t [46]. A second critical feature of multivariate
methods is the use of the NIPALS [47] and related algorithms [35], which
are able to handle small amounts of missing data without resorting to
possibly misleading imputation (guessing) methods. Models are built
with the data that are present, effectively ignoring the missing parts. Only
some of the R packages implement cross-validation and missing value
tolerance. Two of these are pcaMethods [42] and kopls [38].
4.8 Performing PCA on metabolomics
data in R/KNIME
The pcaMethods package contains all required functionality in order to
perform a simple PCA analysis. The input is an internal standard
normalised data set as described in the previous section. A KNIME
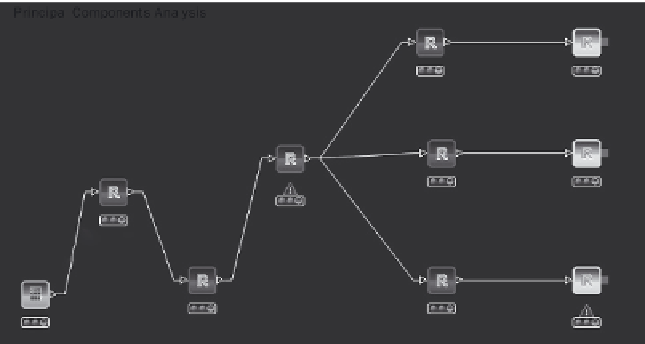
workfl ow to perform this analysis is shown schematically in Figure 4.15
and serves as a useful container for a number of small R scripts.
The PAR Scale node is a wrapper around R package pcaMethods, in
this example, using Pareto scaling. (Pareto scaling is commonly applied
to 'omics data sets and is the division of mean centred variable columns
by the square root of the standard deviation. It up-weights medium scale
features without excessive increases in baseline noise.)
require(pcaMethods)
R<-prep(R, scale = 'pareto',centre = TRUE)
require(pcaMethods)
R<-prep(R, scale = 'pareto',centre = TRUE)
Figure 4.15
PCA analysis using KNIME and R















































Search WWH ::

Custom Search