Biomedical Engineering Reference
In-Depth Information
table. If, for instance, citric acid occurs as both [M+H], [M+Na] adducts
then the workfl ow will add those peaks together. The actual workfl ow as
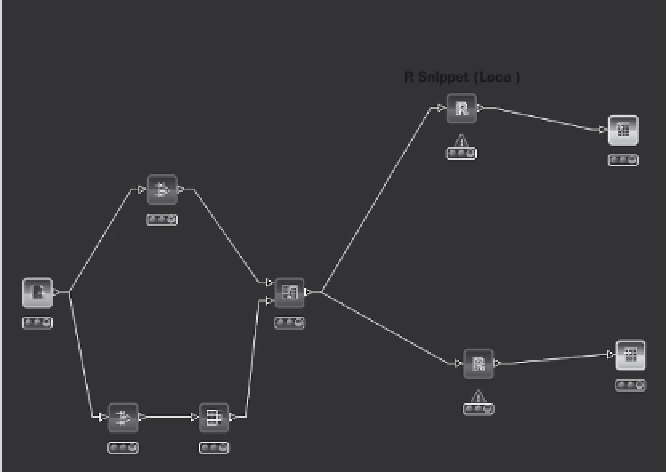
seen in the KNIME GUI is shown in Figure 4.13. This workfl ow reads a
CSV fi le, replaces adduct strings, converts all names to lowercase then
subtotals the spreadsheet. In fact, in a spreadsheet such as Excel, string
replacement and subtotalling are quite involved procedures and with
large data sets the subtotalling may take a long time. In contrast, the
KNIME workfl ow takes seconds to run and will run in a consistent
manner eliminating manual processing errors. The workfl ow is constructed
using various nodes. For example, the String Replacer node replaces any
occurrence of a text (such as [M+H]) in the Name column. The Group By
node groups rows by the Name column, here aggregations such as mean
and sum of row subsets can be performed. The output is a CSV fi le with
only the summed, identifi ed metabolites in alphabetical order.
4.6.2 Internal standard normalisation using
KNIME and R
In a typical metabolomics experiment, it is standard practice to include
isotopic internal standards (such as d4-alanine, d5-phenylalanine) which
Workfl ow to normalise to internal standard or
total signal
Figure 4.14

























Search WWH ::

Custom Search