Biomedical Engineering Reference
In-Depth Information
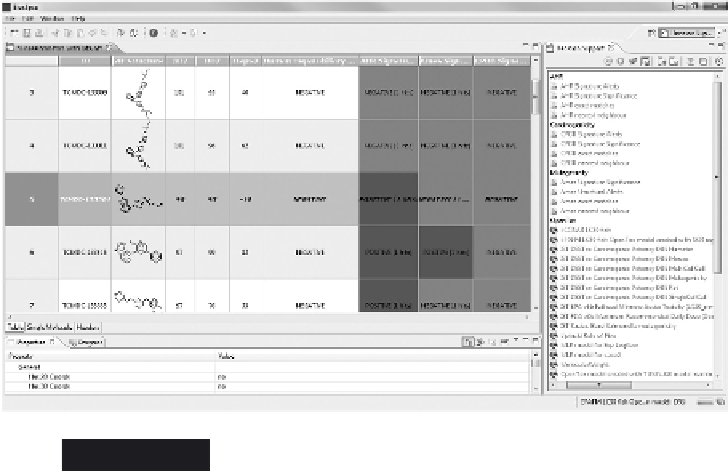
Adding Decision Support columns to the molecule
table. The highlighted compound - TCMDC-133364
- is an interesting candidate as it is highly active
against both strains of
P. falciparum
while being
inactive against human HepG2 cells. Also, the
compound is predicted negative for three out of four
toxicity models included
Figure 2.10
Bioclipse models are negative as well (Figure 2.10). Double-clicking on
the 2D-structure diagram of TCMDC-133364 opens the compound in
the Single Molecule View. We can now change to the Decision Support
perspective and apply the OpenTox models on the selected compound
(Figure 2.11).
The positive AHR Signature Signifi cance can be traced back to
individual atoms in the Decision Support View, allowing for judgment of
the prediction's relevance. The negative prediction for carcinogenicity
by the CPDB model in Bioclipse is confi rmed by the negative predictions
of the OpenTox models 'IST DSSTox Carcinogenic Potency DBS
Hamster', 'IST DSSTox Carcinogenic Potency DBS Mouse' and 'ToxTree:
Benigni/Bossa rules for carcinogenicity and mutagenicity'. Only the
OpenTox models 'IST DSSTox Carcinogenic Potency DBS MultiCellCall'
and 'IST DSSTox Carcinogenic Potency DBS SingleCellCall' are positive.
For mutagenicity we have the negative prediction by Bioclipse's Ames
model, the negative prediction with the OpenTox Benigni/Bossa model



Search WWH ::

Custom Search