Biology Reference
In-Depth Information
denatured
1 chain-enriched chains of this collagen IV that are one to two orders of
magnitude
weaker
than MMP-2, but (b) a
K
m
for intact
a
2 chain-enriched collagen
IV that is one order of magnitude
tighter
than MMP-2 (Gioia et al.
2009
). The
higher affinity of MMP-2 for most protein fibrils evaluated could result from addi-
tional contacts made by two to three of the FnII-like modules (Briknarova et al.
2001
; Gehrmann et al.
2004
; Xu et al.
2009
) (Fig.
6.4
).
The mapping of sites within the basket-shaped FnII-like modules of MMP-2 that
may contact triple-helical and gelatin-like peptides was performed by NMR chemi-
cal shift mapping of single FnII-like modules from MMP-2 (Briknarova et al.
1999
,
2001
) or the complete CBD of MMP-2 (Gehrmann et al.
2004
; Xu et al.
2009
), and
mutagenesis of the CBD (Xu et al.
2009
). Residues identified in at least two of these
studies have been plotted in Fig.
6.4
. These interfacial residues form a bowl in each
module where a tryptophan side chain forms its hydrophobic floor above a two-
stranded
b
-sheet (Fig.
6.5
). Each bowl may be able to accommodate one or perhaps
two bulky and hydrophobic amino acid side chains. Triple helix or gelatin strands
can run horizontally, with only a modest bend, from the active site at left to reach the
binding site in module 3 (blue) at right in Fig.
6.4
. For triple helix or gelatin strands
to run from the active site to the principal binding site in module 2 (green at top in
Fig.
6.4
) or the lesser binding site in module 1 (red in background at right in Fig.
6.4
)
requires considerable bending of the chains. One potential path for gelatin or a triple
helix to traverse from the principal binding site in FnII-like module 2 to the active
site is the channel between catalytic and FnII-like insert domains, in either MMP-
2 or -9 (Fig.
6.4
). A path through this groove is attractive because it passes by the
main binding site in module 2 (top in Fig.
6.4
) and across the exosite proposed to lie
in the catalytic domain to the right of Ca
++
1 (center of Fig.
6.4
and dark red in
a
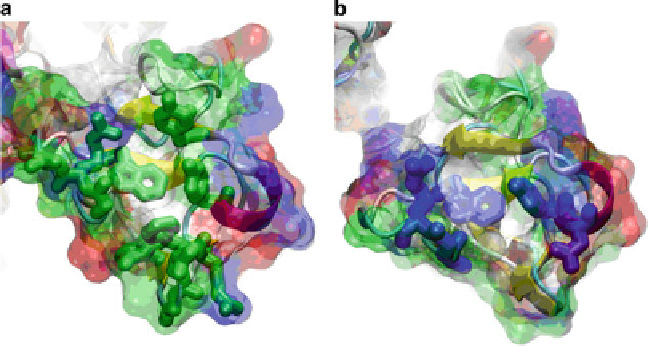
Fig. 6.5 The bowl-like arrangements of residues that were reproducibly found to interact with
gelatin-like or triple-helical peptides (see text) from FnII-like modules 2 and 3 are shown in panels
a and b, respectively. The module 2 residues are plotted with
green side chains
(a) and module 3
with blue side chains (b). The color code of the transparent surface is
green
for polar residues,
white
for hydrophobic residues,
blue
for basic residues, and
red
for acidic residues