Biology Reference
In-Depth Information
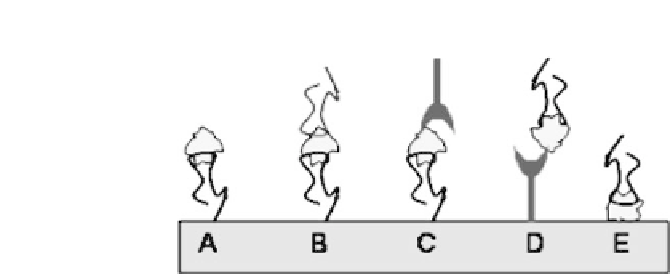
Figure 2.1.
Schematic representation of aptamer-based assays configu-
ration. (A) Direct assay (B) Sandwich assay format with two aptamers (C)
Sandwich assay format with an aptamer used as the primary ligand and an
antibody as the secondary ligand (D) The opposite configuration to case C
(E)Competitive assay.
In a direct assay (Fig. 2.1A) the aptamer is immobilized on the
solid support and the binding of analyte is monitored; a sandwich
assay can be carried out using two aptamers as ligands or combin-
ing an aptamer with an antibody. In this assay format, a capturing
aptamer or antibody is first immobilized on the solid support and
then analyte is added so that the capturing ligand could bind it.
At this point, a detection aptamer or antibody is added and binds
with another site of the target analyte (Fig. 2.1B-D). In addition to
detecting macromolecules, such as proteins, small ligands can also
be bound by aptamers. For this purpose, a competitive assay can
be performed by immobilizing the analyte on the solid support and
then adding to it a solution containing the target analyte and a fixed
and optimizedconcentration of aptamer.
The main differences between the different formats are the
immobilizedspecies(aptamer,antibody,ortargetanalyte),thenum-
ber of experimental steps involved, and in which order the differ-
ent reagents are exposed to the surface. The choice of the format
depends on the molecular size of the analyte, the availability of
reagents, and the cost. The main advantages involved in the use of
a sandwich format are the selectivity and sensitivity of the assay.
When it is possible to perform different assay formats for the detec-
tion of the same target analyte, it is useful to compare the analytical