Information Technology Reference
In-Depth Information
123
31
4
0
Generator
Auto Grid
45
3
5
67
5
67
Generator
2
01
0
1
3
0
1
3
0
2
3
Auto Dock
Auto Dock
Public
Autodock
Vina112
2
2
1
012
012
10
Collector
Collector
Collector.sh
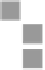
FIGURE 10.4
AutoDock workflow.
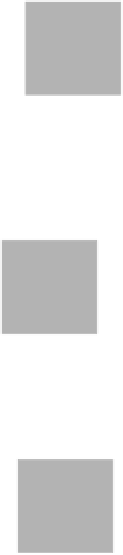
FIGURE 10.5
AutoDock workflow without AutoGrid.
FIGURE 10.6
AutoDock Vina workflow.
computer prior to the execution of the workflow. Although this requires
specific expertise, it also gives much more flexibility to the end user when
preparing the input molecule and the docking space. As a consequence,
this workflow requires PDBQT input files and the output of the AutoGrid
application, and (similar to the previous workflow) it docks a small ligand
molecule on a larger receptor molecule structure in a Monte Carlo simu-
lation using 4.2.3 of the AutoDock docking simulation package. Users of
this workflow are expected to provide a docking parameter file, a ZIP file
containing input files for AutoDock that were generated using version 4.2.3
of AutoGrid and the former docking parameter file, the number of simula-
tions to be carried out, and the number of required results. This workflow is
shown in Figure 10.5.
Finally, the AutoDock Vina workflow performs virtual screening of mole-
cules using version 1.1.2 of AutoDock Vina. It docks a library of small ligands
on a selected receptor molecule. Users of this workflow are expected to pro-
vide a configuration file for AutoDock Vina, an input receptor molecule in
PDBQT file format, a ZIP file containing a number of ligands in PDBQT file
format, the number of simulations to carry out, and the number of required
results. The workflow is shown in Figure 10.6.




























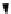













Search WWH ::

Custom Search