Biology Reference
In-Depth Information
Figure 1.5
Potential registries of A
(16-23), KLVFFAED. Favorable
cross-strand pairings for each registry are highlighted in
color where the negatively charged residues E and D side
chains associate with the positively charged K side chain.
This peptide was backbone capped, as was A
β
β
(16-22), with
N-terminal acetyl and C-terminal -NH
.
2
A
B
0.7
0.6
0.5
0.4
0.3
0.2
0.1
0.0
0
5
10 15 20 25 30 35 40 45 50
Time [ms]
C
D
5.8Å
4.2Å
Figure 1.6
(
) Electron micrograph of KLVFFAED peptide assembled
under neutral conditions (identical to A
A
β
(16-22) [23]) for
a week into 15-50 nm bundled sheets. (
)
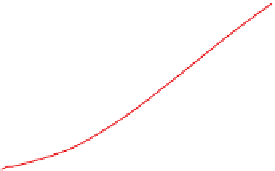
13
C{
15
N} REDOR
of KL[1-
13
C]VFF[
15
N]AED. Solid black line is the calculated
13
B
C{
15
N} REDOR curve for
13
C-
15
N distances shown in (
)
with an N-C-N angle of 156°, consistent with in-register
anti-parallel
C
β
13
C{
15
N}REDOR curve for out-of-
-sheets.
register
β
-sheets (red) and parallel
β
-sheets (dashed line).
(
D
) Model for in-register anti-parallel A
β
(16-23)
β
-sheet
consistent with the NMR distance constraints.
In longer peptide sequences, the sheer number of interactions
could well attenuate the global morphological attenuation of single




































































Search WWH ::

Custom Search