Biology Reference
In-Depth Information
β
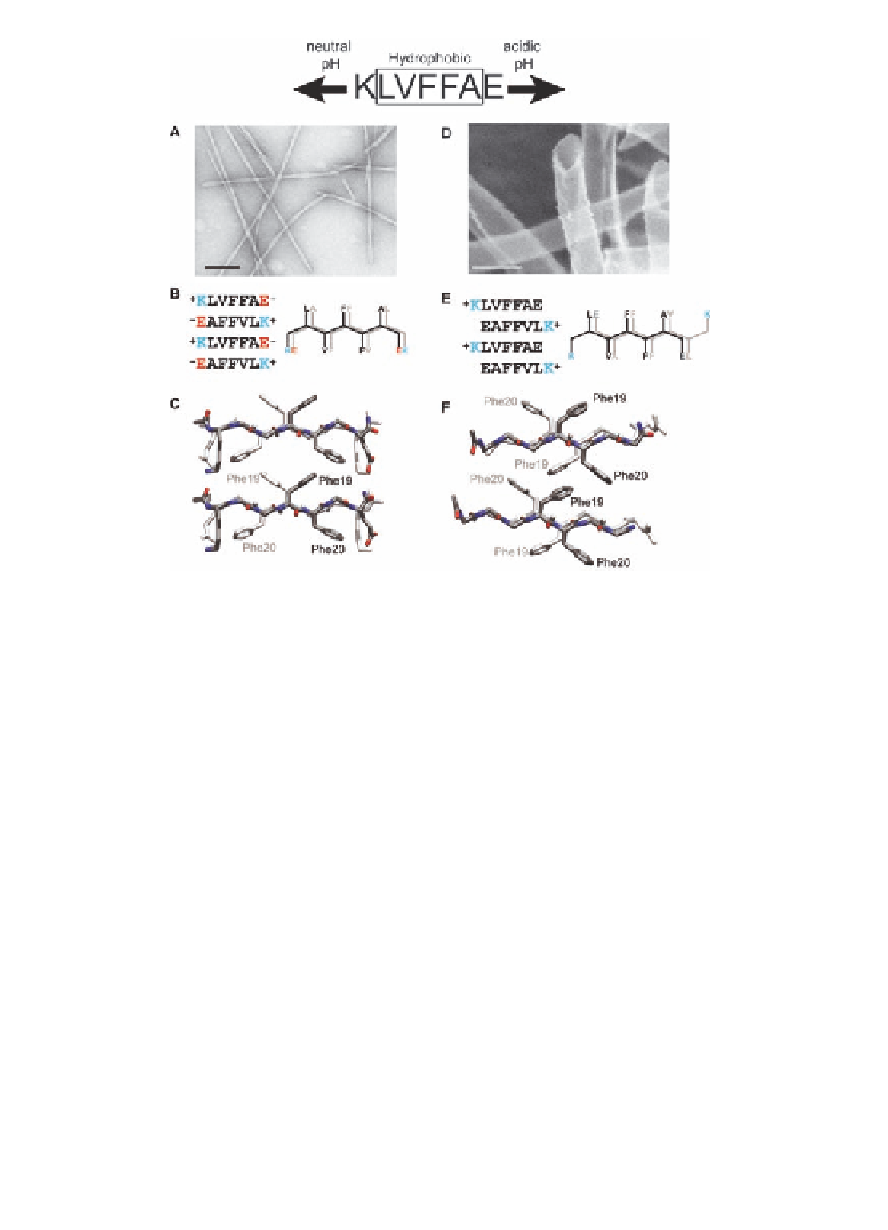
Figure 1.3
The pH-dependent morphology of KLVFFAE, A
(16-22),
fibers (left), and nanotubes (right) [23]. Electron micrograph
images of peptide assembled at (
A
) neutral and (
D)
acidic
conditions. (
) The peptide organization of the fiber
assembly is shown with the Lysine (K) and Glutamic acid (E)
salt bridges directing in-register
B
β
) Protonation
of glutamic acid side chain appears to sufficiently weaken
this electrostatic interaction to allow the packing of the
β
-sheets. (
E
-branched valine against the less bulky alanine to dictate
registry. Structures from molecular dynamics simulations
that match the laminate diffraction distance of ~10 Å for
(
) tubes [23], highlighting the phenylalanine
orientations. Each image consists of four peptides, with the
back H-bonded peptide shaded white. Scale bars in (
C
) fibers and (
F
A
) and
(
D
) are 100 nm.
We will return later to discussions of the diversity of these
structures, but the dynamics of their assembly is most clearly seen
in the time-lapsed images of their assembly. The tube sizes are
below the limit of traditional optical resolution, and initial attempts
to visualize the earliest events associated with the intermolecular
peptide assembly employed fluorescence imaging. It was found
Search WWH ::

Custom Search