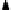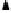Biomedical Engineering Reference
In-Depth Information
Pos.
(kbp)
0.3
1.547
1.469
1.391
1.313
1.235
1.157
1.079
1.001
0.923
0.845
0.767
0.689
0.611
0.533
0.455
0.377
0.299
0.221
0.413
0.065
0
50
60
70
80
90
Temperature (
°
C)
FIGURE 1.47
MELTSIM simulated first-derivative melting plot of the fraction theta of the
D. discoideum
diphosphatase gene
melted (left-hand
Y
-axis) as a function of temperature presented as the solid line. The right-hand
Y
-axis shows
the positions along the diphosphatase gene, as does the exon and intron locations described by the labeled ver-
tical arrows on the left-hand portion of the figure. The positional melting simulation results along the diphos-
phatase gene are indicated by the temperatures of the leftward bolded edges of the shaded areas that correspond
to the melting at that gene position segment. The bolded melting regions can be seen to correspond to the posi-
tions of peaks in the total gene melting description depicted by the solid line. Reprinted from Marx, K.A., Assil,
I.Q., Bizzaro, J.W., Blake, R.D. (1998). Comparison of Experimental to MELTSIM Calculated DNA Melting of the
(A
T) Rich
Dictyostelium discoideum
Genome: Denaturation Maps Distinguish Exons from Introns.
J. Biomol.
Struct. Dynamics
16:329-340. With permission from Adenine Press.
array and allowed to hybridize to their immobilized complementary nucleic acid
sequence, followed by washing to remove unhybridized sequences. The fluorescence
intensities of all hybridized spots, reflecting the in vivo mRNA levels, are then deter-
mined. This type of large-scale gene product analysis is critically important for under-
standing the molecular basis of serious and complex biomedical conditions, such as
cancer, where interconnected gene networks can produce nonlinear molecular effects
characteristic of the biomedical state being analyzed. Increasingly, the nucleic acids
being incorporated into microarray spots can be quite long, hundreds of base pairs, con-
siderably longer than the thermodynamic block melting size found in typical DNAs.
Therefore, complex multiblock melting behavior similar to that being simulated by
MELTSIM will increasingly occur within hybrids formed on the microarray chips. For
reasons such as this, we believe that our demonstration of the utility of MELTSIM to
accurately simulate long DNA sequence melting in a position-dependent fashion along
the sequence will facilitate the rational incorporation of long DNAs into smart biosen-
sor designs.