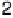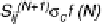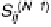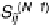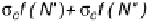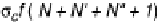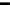Biomedical Engineering Reference
In-Depth Information
duplexes. DNA melting carried out experimentally was one of the early biophysical tools
used to characterize genomic DNA (148), before the development of the modern tech-
niques of molecular biology such as restriction enzyme analysis, cloning, and fast
sequence analysis. With the advent of solid-phase DNA synthesis and PCR technology in
the 1980s, the use of short DNA single-stranded sequences, ranging from around 10-300 nt
in size, has rapidly increased. These short DNA sequences are in widespread use in both
research and diagnostic applications as biosensor probes. They can be used to detect the
occurrence and often the concentration of DNA from an infectious agent or abnormal tis-
sue state found in cells or patient samples in clinical settings (149). The thermodynamic
stability, represented by the
T
m
of the hybrids formed in these assays as well as that of the
short DNA duplexes synthesized in the ubiquitous PCR reaction, are important quantities
for investigators to estimate or to calculate for use in experimental protocols. Typically,
these
T
m
values are determined experimentally or are estimated using nonrigorous rules
employing the simple base composition of the sequence as input in the estimation.
It has been known for some time that experimental DNA melting could be simulated
using a statistical mechanical approach. The Poland-Scheraga algorithm is a methodology
that was developed to describe the energetically cooperative process of equilibrium DNA
melting. This approach uses a two-state model where a series of fundamental equilibrium
energetic states, shown in Figure 1.46, represent the complex process of long DNA
sequences of different local base composition (melting blocks), melting as thermodynamic
blocks at different
T
m
values (150). In the program MELTSIM, this algorithm has been
implemented and parameterized for long DNA sequences with the experimentally deter-
mined thermodynamic quantities for all 16 [(4 possible bases)
2
] of the possible DNA din-
ucleotide pairs (151).
At the Center for Intelligent Biomaterials, we have utilized MELTSIM to study a num-
ber of features of the melting of long DNA sequences. In the broadest genomic survey
conducted to date, we used MELTSIM to study the melting behavior of whole chromo-
somes and populations of long single gene-containing sequences from 28 eukaryotic
organisms covering the entire (G
C)% composition range from about 20 to 60% (152).
For over 8000 sequences in these organisms, we demonstrated a high correlation
between each sequence's calculated
T
m
value and its fraction (G
C) base composition.
Through a series of melting simulations of control sequences created as a result of
Case:
I
II
III
IV
V
N'
N"
N'
N'
N
N
Process:
N
N
N
Boundaries:
+2
+1
0
Equilibrium
constant
S
ij
N
S
ij
(
N+1
)
:
FIGURE 1.46
Schematic representation of five of the six cases of thermal melting processes of double helical DNA, with
changes in the numbers of boundaries and equilibrium constants indicated. Reprinted from Blake, R.D., Bizzaro,
J.W., Blake, J.D., Day, G.R., Delcourt, S.G., Knowles, J., Marx, K.A., SantaLucia, Jr., J. (1999). Statistical Mechanical
Simulation of Polymeric DNA Melting With MELTSIM.
Bioinformatics
15:370-375. With permission of Oxford
University Press.