Biomedical Engineering Reference
In-Depth Information
5
′
C
3
′
Constant
DNA library
Random sequence
Step 1
Transcription
Step 2
5
3
′
RNA library
Target
Folded
aptamers
Mixing
Step 3
Step 7 (10-12 x)
Partition
Discard
Enrich
Step 4
Reverse
transcription
5
3
Step 5
3
5
5
3
Step 6
3
5
PCR
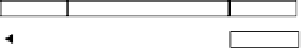
FIGURE 20.6
The SELEX Protocol. (Modified from Sampson T. 2003. Aptamers and SELEX: The technology.
World Patent
Information
25: 123-129) (see text for details).
Referring to Figure 20.6, Steps 1 and 2 involve the preparation of the aptamer
library/candidate mixture, as detailed above. Note that the library would generally con-
sist of double-stranded DNA, which needs to be either transcribed (for RNA selection) or
strand-separated (for ss DNA selection), to be in a suitable form for selection (54).
In Step 3, the target and the aptamers are brought together under favorable binding con-
ditions, where the aptamers with the highest affinity will bind the target. These aptamers
are then partitioned from the aptamers with lower affinity in Step 4. This step can be per-
formed by attaching the aptamers to a solid phase support, such as Sepharose, and specif-
ically eluting the desired aptamers after binding has taken place. Immobilizing the
aptamers, however, is likely to affect the binding interactions between the aptamer and the
target. The same would be true if the target were immobilized instead of the aptamer (53).
Alternatively, the aptamer and target could be allowed to interact freely in solution, after
which the target-aptamer complex could be recovered by filtration through nitrocellulose
(54). This method is commonly used, although it is important to note that it is only appli-
cable when the target molecule is a protein. A negative selection step is also frequently